| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-09-08 17:50:22 -0600 |
|---|
| Update Date | 2015-09-14 16:46:02 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
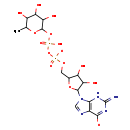
| Name: | GDP-β-L-fucose |
|---|
| Description | GDP-beta-L-fucose is a sugar nucleotide and a readily available source of fucose. The monosaccharide plays several important metabolic roles in complex carbohydrates and in glycoproteins. Fucosylated oligosaccharides are involved in cell-cell recognition, selectin-mediated leukocyte-endothelial adhesion, and mouse embryogenesis. Fucose is made available during the synthesis of fucosylated glycolipids, oligosaccharides, and glycoproteins via a sugar nucleotide intermediate, specifically GDP-beta-L-fucose.GDP-beta-L-fucose pyrophosphorylase (GFPP, E. C. 2.7.7.30) catalyzes the reversible condensation of guanosine triphosphate and beta-L-fucose-1-phosphate to form the nucleotide-sugar GDP-beta-L-fucose. |
|---|
| Structure | |
|---|
| Synonyms: | Not Available |
|---|
| Chemical Formula: | C16H23N5O15P2 |
|---|
| Weight: | Average: 587.329
Monoisotopic: 587.067686216 |
|---|
| InChI Key: | LQEBEXMHBLQMDB-UHFFFAOYSA-L |
|---|
| InChI: | InChI=1S/C16H25N5O15P2/c1-4-7(22)9(24)11(26)15(33-4)35-38(30,31)36-37(28,29)32-2-5-8(23)10(25)14(34-5)21-3-18-6-12(21)19-16(17)20-13(6)27/h3-5,7-11,14-15,22-26H,2H2,1H3,(H,28,29)(H,30,31)(H3,17,19,20,27)/p-2 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | [5-(2-amino-6-oxo-6,9-dihydro-3H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(3,4,5-trihydroxy-6-methyloxan-2-yl phosphonato)oxy]phosphonate |
|---|
| Traditional IUPAC Name: | [5-(2-amino-6-oxo-3H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (3,4,5-trihydroxy-6-methyloxan-2-yl phosphonato)oxyphosphonate |
|---|
| SMILES: | CC1OC(OP([O-])(=O)OP([O-])(=O)OCC2OC(C(O)C2O)N2C=NC3=C2NC(N)=NC3=O)C(O)C(O)C1O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine nucleotide sugars |
|---|
| Direct Parent | Purine nucleotide sugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine nucleotide sugar
- Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-oxopurine
- Hypoxanthine
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Purine
- Pyrimidone
- Aminopyrimidine
- Pyrimidine
- Alkyl phosphate
- Phosphoric acid ester
- Oxane
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Vinylogous amide
- Azole
- Imidazole
- Oxolane
- Heteroaromatic compound
- Secondary alcohol
- 1,2-diol
- Polyol
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Hydrocarbon derivative
- Organic oxide
- Organic oxygen compound
- Organic nitrogen compound
- Primary amine
- Alcohol
- Organooxygen compound
- Organonitrogen compound
- Amine
- Organic anion
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | Amino sugar and nucleotide sugar metabolism II | PW000887 | 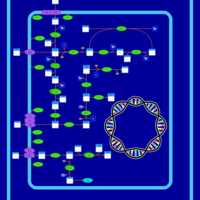 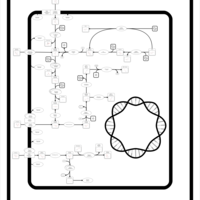 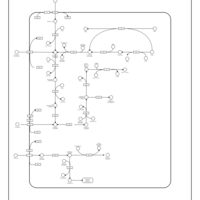 |
|
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | 20705994 | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|