| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-09-08 17:50:21 -0600 |
|---|
| Update Date | 2015-09-14 16:46:14 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
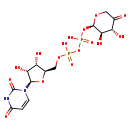
| Name: | UDP-β-L-threo-pentapyranos-4-ulose |
|---|
| Description | UDP-beta-L-threo-pentapyranos-4-ulose is an intermediate in the polymixin resistance pathway. It is a substrate for the enzyme UDP-4-amino-4-deoxy-L-arabinose aminotransferase which catalyzes the reaction UDP-4-amino-4-deoxy-beta-L-arabinopyranose + 2-oxoglutarate = UDP-beta-L-threo-pentapyranos-4-ulose + L-glutamate. Some Gram-negative bacteria, specifically Salmonella typhimurium and Escherichia coli, can become resistant to polymyxin by the modification of their lipid A structure via the attachment of 4-amino-4-deoxy-L-arabinopyranose (L-Ara4N) groups to one or more phosphate groups. This addition causes an absolute increase in lipid A charge, thus lowering the affinity of positively charged polymyxins. |
|---|
| Structure | |
|---|
| Synonyms: | | Value | Source |
|---|
| UDP-4''-Ketopentose | ChEBI | | UDP-beta-L-Threo-pentapyranos-4-ulose | ChEBI | | UDP-L-Ara4O | ChEBI | | Uridine 5'-(beta-L-threo-pentapyranosyl-4''-ulose diphosphate) | ChEBI | | Uridine 5'-diphospho-beta-(L-threo-pentapyranosyl-4''-ulose) | ChEBI | | UDP-4-Keto-D-xylose | Kegg | | UDP-b-L-Threo-pentapyranos-4-ulose | Generator | | Uridine 5'-(b-L-threo-pentapyranosyl-4''-ulose diphosphate) | Generator | | Uridine 5'-(b-L-threo-pentapyranosyl-4''-ulose diphosphoric acid) | Generator | | Uridine 5'-(beta-L-threo-pentapyranosyl-4''-ulose diphosphoric acid) | Generator | | Uridine 5'-(β-L-threo-pentapyranosyl-4''-ulose diphosphate) | Generator | | Uridine 5'-(β-L-threo-pentapyranosyl-4''-ulose diphosphoric acid) | Generator | | Uridine 5'-diphospho-b-(L-threo-pentapyranosyl-4''-ulose) | Generator | | Uridine 5'-diphospho-β-(L-threo-pentapyranosyl-4''-ulose) | Generator | | UDP-b-L-threo-Pentopyranos-4-ulose | Generator | | UDP-β-L-threo-pentopyranos-4-ulose | Generator | | UDP-β-L-threo-pentapyranos-4-ulose | Generator |
|
|---|
| Chemical Formula: | C14H20N2O16P2 |
|---|
| Weight: | Average: 534.26
Monoisotopic: 534.028806568 |
|---|
| InChI Key: | URJZIQLTPCJVMW-QNSCKLTRSA-N |
|---|
| InChI: | InChI=1S/C14H20N2O16P2/c17-5-3-28-13(11(22)8(5)19)31-34(26,27)32-33(24,25)29-4-6-9(20)10(21)12(30-6)16-2-1-7(18)15-14(16)23/h1-2,6,8-13,19-22H,3-4H2,(H,24,25)(H,26,27)(H,15,18,23)/t6-,8+,9-,10-,11-,12-,13-/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | [({[(2R,3R,4R)-3,4-dihydroxy-5-oxooxan-2-yl]oxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy})phosphinic acid |
|---|
| Traditional IUPAC Name: | udp-L-ara4O |
|---|
| SMILES: | O[C@@H]1[C@@H](COP(O)(=O)OP(O)(=O)O[C@H]2OCC(=O)[C@H](O)[C@H]2O)O[C@H]([C@@H]1O)N1C=CC(=O)NC1=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine ribonucleoside diphosphates. These are pyrimidine ribonucleotides with diphosphate group linked to the ribose moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine ribonucleotides |
|---|
| Direct Parent | Pyrimidine ribonucleoside diphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine ribonucleoside diphosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Organic pyrophosphate
- Pyrimidone
- Monoalkyl phosphate
- Hydropyrimidine
- Monosaccharide
- Organic phosphoric acid derivative
- Oxane
- Phosphoric acid ester
- Pyrimidine
- Alkyl phosphate
- Vinylogous amide
- Heteroaromatic compound
- Tetrahydrofuran
- Cyclic ketone
- Ketone
- Urea
- Secondary alcohol
- Lactam
- Azacycle
- Oxacycle
- Organoheterocyclic compound
- Alcohol
- Organopnictogen compound
- Carbonyl group
- Organic oxide
- Organonitrogen compound
- Organooxygen compound
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxygen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | Amino sugar and nucleotide sugar metabolism III | PW000895 |  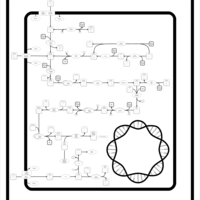 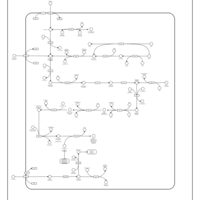 | | polymyxin resistance | PW002052 | 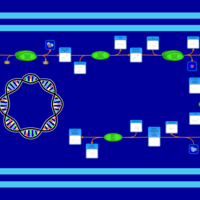 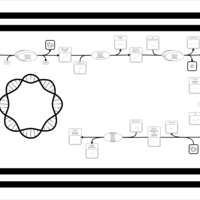 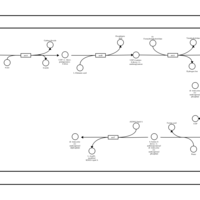 |
|
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | 47028 | | HMDB ID | Not Available | | Pubchem Compound ID | 17756767 | | Kegg ID | C16155 | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|