| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-09-08 17:50:19 -0600 |
|---|
| Update Date | 2015-09-14 16:46:11 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | S-ribosyl-L-homocysteine |
|---|
| Description | S-Ribosyl-L-homocysteine is a member of the chemical class known as Alpha Amino Acids and Derivatives. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon). AI-2 is spontaneously derived from 4,5-dihydroxy-2,3-pentanedione that, along with homocysteine, is produced by cleavage of S-adenosylhomocysteine (SAH) and S-ribosylhomocysteine by the Pfs and LuxS enzymes. (PMID 16885435) |
|---|
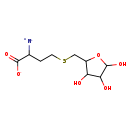
| Structure | |
|---|
| Synonyms: | Not Available |
|---|
| Chemical Formula: | C9H14NO6S |
|---|
| Weight: | Average: 264.27
Monoisotopic: 264.054183349 |
|---|
| InChI Key: | QUDMRZABJLFLCP-UHFFFAOYSA-M |
|---|
| InChI: | InChI=1S/C9H15NO6S/c10-4(8(13)14)1-2-17-3-5-6(11)7(12)9(15)16-5/h4-7,9,11-12,15H,1-3H2,(H,13,14)/q+1/p-1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (1-carboxylato-3-{[(3,4,5-trihydroxyoxolan-2-yl)methyl]sulfanyl}propyl)azaniumyl |
|---|
| Traditional IUPAC Name: | (1-carboxylato-3-{[(3,4,5-trihydroxyoxolan-2-yl)methyl]sulfanyl}propyl)ammonio |
|---|
| SMILES: | [N+]C(CCSCC1OC(O)C(O)C1O)C([O-])=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-amino acid or derivatives
- Pentose monosaccharide
- Heterocyclic fatty acid
- Hydroxy fatty acid
- Thia fatty acid
- Fatty acyl
- Fatty acid
- Tetrahydrofuran
- 1,2-diol
- Carboxylic acid salt
- Hemiacetal
- Secondary alcohol
- Dialkylthioether
- Sulfenyl compound
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Polyol
- Oxacycle
- Organoheterocyclic compound
- Thioether
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Organonitrogen compound
- Organosulfur compound
- Hydrocarbon derivative
- Organic zwitterion
- Organic nitrogen compound
- Alcohol
- Organic salt
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 0 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | Not Available | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|