| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-09-08 17:50:09 -0600 |
|---|
| Update Date | 2016-09-13 16:35:45 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
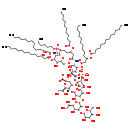
| Name: | glucosyl-(heptosyl)2-Kdo2-lipid A |
|---|
| Description | Glucosyl-(heptosyl)2-Kdo2-lipid A is an intermediate in lipid A-core biosynthesis pathway in E.coli. It is a substrate for the enzyme lipopolysaccharide core heptose (I) kinase which catalyzes the reaction glucosyl-(heptosyl)2-Kdo2-lipid A + ATP -> glucosyl-(heptosyl)2-Kdo2-lipid A-phosphate + ADP + H+. It is also a product for enzyme lipopolysaccharide glucosyltransferase I which catalyzes reaction UDP-alpha-D-glucose + (heptosyl)2-Kdo2-lipid A -> glucosyl-(heptosyl)2-Kdo2-lipid A + UDP + H+ (BioCyc compound: CPD0-932). |
|---|
| Structure | |
|---|
| Synonyms: | | Value | Source |
|---|
| D-GLC-(1->3)-L-alpha-D-hep-(1->3)-L-alpha-D-hep-(1->5)-[alpha-kdo-(2->4)]-alpha-kdo-(2->6)-lipid a hexaanion | ChEBI | | D-GLC-(1->3)-L-alpha-D-hep-(1->3)-L-alpha-D-hep-(1->5)-[alpha-kdo-(2->4)]-alpha-kdo-(2->6)-lipid a(6-) | ChEBI | | Glucosyl-(heptosyl)2-(kdo)2-lipid a | ChEBI | | Glucosyl-(heptosyl)2-(kdo)2-lipid a hexaanion | ChEBI |
|
|---|
| Chemical Formula: | |
|---|
| Weight: | Average: Not Available
Monoisotopic: Not Available |
|---|
| InChI Key: | Not Available |
|---|
| InChI: | Not Available |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2R,4R,5R,6R)-2-{[(2R,4R,5R,6R)-2-carboxylato-6-[(1R)-1,2-dihydroxyethyl]-5-{[(2R,3S,4S,5R,6R)-6-[(1S)-1,2-dihydroxyethyl]-4-{[(2R,3S,4S,5R,6R)-6-[(1S)-1,2-dihydroxyethyl]-3,5-dihydroxy-4-{[(3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}oxan-2-yl]oxy}-3,5-dihydroxyoxan-2-yl]oxy}-2-{[(2R,3S,4R,5R,6R)-5-[(3R)-3-(dodecanoyloxy)tetradecanamido]-6-{[(2R,3S,4R,5R,6R)-3-hydroxy-5-[(3R)-3-hydroxytetradecanamido]-4-{[(3R)-3-hydroxytetradecanoyl]oxy}-6-(phosphonatooxy)oxan-2-yl]methoxy}-3-(phosphonatooxy)-4-{[(3R)-3-(tetradecanoyloxy)tetradecanoyl]oxy}oxan-2-yl]methoxy}oxan-4-yl]oxy}-6-[(1R)-1,2-dihydroxyethyl]-4,5-dihydroxyoxane-2-carboxylate |
|---|
| Traditional IUPAC Name: | (2R,4R,5R,6R)-2-{[(2R,4R,5R,6R)-2-carboxylato-6-[(1R)-1,2-dihydroxyethyl]-5-{[(2R,3S,4S,5R,6R)-6-[(1S)-1,2-dihydroxyethyl]-4-{[(2R,3S,4S,5R,6R)-6-[(1S)-1,2-dihydroxyethyl]-3,5-dihydroxy-4-{[(3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}oxan-2-yl]oxy}-3,5-dihydroxyoxan-2-yl]oxy}-2-{[(2R,3S,4R,5R,6R)-5-[(3R)-3-(dodecanoyloxy)tetradecanamido]-6-{[(2R,3S,4R,5R,6R)-3-hydroxy-5-[(3R)-3-hydroxytetradecanamido]-4-{[(3R)-3-hydroxytetradecanoyl]oxy}-6-(phosphonatooxy)oxan-2-yl]methoxy}-3-(phosphonatooxy)-4-{[(3R)-3-(tetradecanoyloxy)tetradecanoyl]oxy}oxan-2-yl]methoxy}oxan-4-yl]oxy}-6-[(1R)-1,2-dihydroxyethyl]-4,5-dihydroxyoxane-2-carboxylate |
|---|
| SMILES: | Not Available |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as acylaminosugars. These are organic compounds containing a sugar linked to a chain through N-acyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Acylaminosugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Oligosaccharide phosphate
- Oligosaccharide
- Hexacarboxylic acid or derivatives
- Acylaminosugar
- Saccharolipid
- Hexose phosphate
- N-acyl-alpha-hexosamine
- Fatty acyl glycoside
- C-glucuronide
- Alkyl glycoside
- O-glycosyl compound
- C-glycosyl compound
- Glycosyl compound
- Fatty acid ester
- Ketal
- Beta-hydroxy acid
- Pyran
- Fatty acyl
- Phosphoric acid ester
- Oxane
- Hydroxy acid
- Organic phosphoric acid derivative
- N-acyl-amine
- Fatty amide
- Alkyl phosphate
- Carboxamide group
- Carboxylic acid ester
- Carboxylic acid salt
- Secondary carboxylic acid amide
- Secondary alcohol
- Organoheterocyclic compound
- Carboxylic acid
- Carboxylic acid derivative
- Acetal
- Oxacycle
- Polyol
- Alcohol
- Organic nitrogen compound
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Primary alcohol
- Organonitrogen compound
- Organic anion
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -6 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Membrane |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | Lipopolysaccharide biosynthesis | PW000831 | 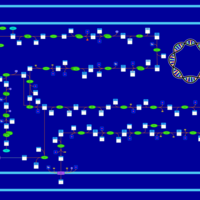 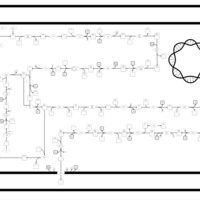 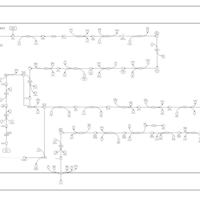 | | lipopolysaccharide biosynthesis II | PW001905 |  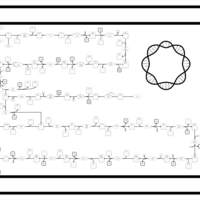  | | lipopolysaccharide biosynthesis III | PW002059 | 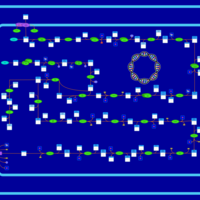 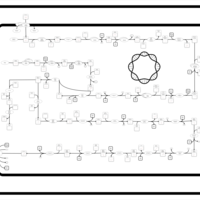 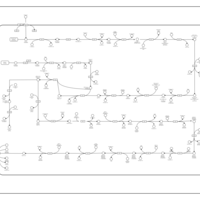 |
|
|---|
| KEGG Pathways: | - Lipopolysaccharide biosynthesis ec00540
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | Not Available |
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | 51351771 | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|