| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-06-08 15:59:17 -0600 |
|---|
| Update Date | 2015-08-05 16:22:05 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Secondary alcohol |
|---|
| Description | A secondary alcohol is a compound in which a hydroxy group, ‒OH, is attached to a saturated carbon atom which has two other carbon atoms attached to it. |
|---|
| Structure | |
|---|
| Synonyms: | - (S)-3'-((2-amino-3-(4-Methoxyphenyl)-1-oxopropyl)amino)-3'-deoxy-N,N-dimethyladenosine
- 3'-(L-a-amino-P-methoxyhydrocinnamamido)-3'-Deoxy-N,N-dimethyladenosine
- 3'-(L-alpha-amino-P-methoxyhydrocinnamamido)-3'-Deoxy-N,N-dimethyladenosine
- 3'-(L-α-amino-P-methoxyhydrocinnamamido)-3'-deoxy-N,N-dimethyladenosine
- 3'-[[(2S)-2-amino-3-(4-Methoxyphenyl)-1-oxopropyl]amino]-3'-deoxy-N,N-diemthyladenosine
- 9-{3-deoxy-3-[(O-methyl-L-tyrosyl)amino]-b-D-xylofuranosyl}-N,N-dimethyl-9H-purin-6-amine
- 9-{3-deoxy-3-[(O-methyl-L-tyrosyl)amino]-beta-D-xylofuranosyl}-N,N-dimethyl-9H-purin-6-amine
- 9-{3-deoxy-3-[(O-methyl-L-tyrosyl)amino]-β-D-xylofuranosyl}-N,N-dimethyl-9H-purin-6-amine
- Achromycin
- Puromicina
- Puromycine
- Puromycinum
|
|---|
| Chemical Formula: | C22H29N7O5 |
|---|
| Weight: | Average: 471.5096
Monoisotopic: 471.223017073 |
|---|
| InChI Key: | RXWNCPJZOCPEPQ-NVWDDTSBSA-N |
|---|
| InChI: | InChI=1S/C22H29N7O5/c1-28(2)19-17-20(25-10-24-19)29(11-26-17)22-18(31)16(15(9-30)34-22)27-21(32)14(23)8-12-4-6-13(33-3)7-5-12/h4-7,10-11,14-16,18,22,30-31H,8-9,23H2,1-3H3,(H,27,32)/t14-,15+,16+,18+,22+/m0/s1 |
|---|
| CAS number: | Not Available |
|---|
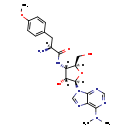
| IUPAC Name: | (2S)-2-amino-N-[(2S,3S,4R,5R)-5-[6-(dimethylamino)-9H-purin-9-yl]-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl]-3-(4-methoxyphenyl)propanamide |
|---|
| Traditional IUPAC Name: | puromycin |
|---|
| SMILES: | [H][C@](N)(CC1=CC=C(OC)C=C1)C(=O)N[C@]1([H])[C@@]([H])(CO)O[C@@]([H])(N2C=NC3=C(N=CN=C23)N(C)C)[C@]1([H])O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine 3'-deoxyribonucleosides. Purine 3'-deoxyribonucleosides are compounds consisting of a purine linked to a ribose which lacks a hydroxyl group at position 3. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleosides |
|---|
| Sub Class | Purine 3'-deoxyribonucleosides |
|---|
| Direct Parent | Purine 3'-deoxyribonucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine 3'-deoxyribonucleoside
- Phenylalanine or derivatives
- Alpha-amino acid amide
- N-glycosyl compound
- Glycosyl compound
- 6-alkylaminopurine
- 6-aminopurine
- Alpha-amino acid or derivatives
- Pentose monosaccharide
- Amphetamine or derivatives
- Imidazopyrimidine
- Purine
- Phenoxy compound
- Phenol ether
- Methoxybenzene
- Anisole
- Dialkylarylamine
- Aralkylamine
- Aminopyrimidine
- Alkyl aryl ether
- Monosaccharide
- Fatty acyl
- Imidolactam
- Benzenoid
- N-substituted imidazole
- Fatty amide
- Monocyclic benzene moiety
- Pyrimidine
- Heteroaromatic compound
- Imidazole
- Azole
- Tetrahydrofuran
- Amino acid or derivatives
- Secondary carboxylic acid amide
- Carboxamide group
- Secondary alcohol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Carboxylic acid derivative
- Ether
- Carbonyl group
- Primary alcohol
- Primary amine
- Organic oxygen compound
- Alcohol
- Organic nitrogen compound
- Hydrocarbon derivative
- Amine
- Organooxygen compound
- Organopnictogen compound
- Organic oxide
- Organonitrogen compound
- Primary aliphatic amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | - 1- and 2-Methylnaphthalene degradation ec00624
- 3-Chloroacrylic acid degradation ec00641
- Butanoate metabolism ec00650
- Chloroalkane and chloroalkene degradation ec00625
- Drug metabolism - cytochrome P450 ec00982
- Fatty acid metabolism ec00071
- Glycine, serine and threonine metabolism ec00260
- Glycolysis / Gluconeogenesis ec00010
- Metabolic pathways eco01100
- Metabolism of xenobiotics by cytochrome P450 ec00980
- Microbial metabolism in diverse environments ec01120
- Naphthalene degradation ec00626
- Pyruvate metabolism ec00620
- Retinol metabolism ec00830
- Tyrosine metabolism ec00350
- alpha-Linolenic acid metabolism ec00592
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_4_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-00di-0000900000-f82e7526c7ebd78049d9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03k9-0900500000-0d48aed0c9497b5ab8f3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03di-0900000000-af8be24075f803a58153 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03di-0900000000-571b6483dc540c02d098 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-03di-0900000000-08775f201fb11e558b53 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-00di-0000900000-6133e2c60106f3437756 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-05fr-0005900000-a760a4943250cd5405b9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0a4i-0809000000-eb619283ee6c689551e3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0w29-0900000000-ca39c6d95b7bd8fba81b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0udi-0900000000-253fadebaa39d03e1cd1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , positive | splash10-0a4i-0219000000-6595a051c26ce2ee007b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900300000-347a5da10b8c2c85e6c0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dj-0900000000-4a2ce92d6902f4ad8eaa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dj-1900000000-039465e4b89c78bd0b11 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0229-0520900000-37dea53aac093ce884f6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0910100000-f60802f57acd3074b4c1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03xs-1900000000-d2fbe55b0731a8cf001f | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|