| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-06-08 15:26:57 -0600 |
|---|
| Update Date | 2015-07-23 12:07:28 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Aromatic oxo acid |
|---|
| Description | |
|---|
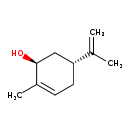
| Structure | |
|---|
| Synonyms: | - (-)-(4R,6S)-trans-Carveol
- Carveol
|
|---|
| Chemical Formula: | C10H16O |
|---|
| Weight: | Average: 152.2334
Monoisotopic: 152.120115134 |
|---|
| InChI Key: | BAVONGHXFVOKBV-ZJUUUORDSA-N |
|---|
| InChI: | InChI=1S/C10H16O/c1-7(2)9-5-4-8(3)10(11)6-9/h4,9-11H,1,5-6H2,2-3H3/t9-,10+/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (1S,5R)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ol |
|---|
| Traditional IUPAC Name: | (-)-trans-carveol |
|---|
| SMILES: | CC(=C)[C@@H]1CC=C(C)[C@@H](O)C1 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as menthane monoterpenoids. These are monoterpenoids with a structure based on the o-, m-, or p-menthane backbone. P-menthane consists of the cyclohexane ring with a methyl group and a (2-methyl)-propyl group at the 1 and 4 ring position, respectively. The o- and m- menthanes are much rarer, and presumably arise by alkyl migration of p-menthanes. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Monoterpenoids |
|---|
| Direct Parent | Menthane monoterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - P-menthane monoterpenoid
- Monocyclic monoterpenoid
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 0 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | - Cysteine and methionine metabolism ec00270
- Isoquinoline alkaloid biosynthesis ec00950
- Lysine biosynthesis ec00300
- Metabolic pathways eco01100
- Novobiocin biosynthesis ec00401
- Phenylalanine metabolism ec00360
- Phenylalanine, tyrosine and tryptophan biosynthesis ec00400
- Tropane, piperidine and pyridine alkaloid biosynthesis ec00960
- Tyrosine metabolism ec00350
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-05nf-9700000000-a75055e28480ba00b820 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0a5c-9300000000-0497c064b7fcb0223af5 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-05nf-9700000000-a75055e28480ba00b820 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0a5c-9300000000-0497c064b7fcb0223af5 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-05nf-9700000000-a75055e28480ba00b820 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0a5c-9300000000-0497c064b7fcb0223af5 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-05nf-9700000000-a75055e28480ba00b820 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0a5c-9300000000-0497c064b7fcb0223af5 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0040-9300000000-900d20824e13ff7b371a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-059l-9420000000-c8fe9a59dae7a88e5d86 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f79-1900000000-6cbdaa00e43b6a5f1466 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0f79-7900000000-b55dc6268bf1d9facfab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-1000-9200000000-82efaa1d3034c0a127ad | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-63662b96265d54ab95ac | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0900000000-fb53bb25a46fab88d48a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0frl-6900000000-b30096dc2484f9341f67 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-c751b1e6b18ebbd8226b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ue9-0900000000-ff7b348d24f4a9fae768 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0159-5900000000-32379e3babd1cfac9d58 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000l-7900000000-14152f364bee2002ab86 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-002f-9100000000-c417dcb5d6ff023a5c44 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-33b66b0d611b3c8dca5f | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|