| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-06-04 16:05:06 -0600 |
|---|
| Update Date | 2015-09-17 15:42:04 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | D-Tagaturonate |
|---|
| Description | Conjugate base of D-tagaturonic acid |
|---|
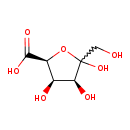
| Structure | |
|---|
| Synonyms: | |
|---|
| Chemical Formula: | C6H9O7 |
|---|
| Weight: | Average: 193.132
Monoisotopic: 193.03537621 |
|---|
| InChI Key: | IZSRJDGCGRAUAR-UHFFFAOYSA-M |
|---|
| InChI: | InChI=1S/C6H10O7/c7-1-2(8)3(9)4(10)5(11)6(12)13/h3-5,7,9-11H,1H2,(H,12,13)/p-1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2S,3R,4S)-3,4,5-trihydroxy-5-(hydroxymethyl)oxolane-2-carboxylic acid |
|---|
| Traditional IUPAC Name: | (2S,3R,4S)-3,4,5-trihydroxy-5-(hydroxymethyl)oxolane-2-carboxylic acid |
|---|
| SMILES: | OCC(=O)C(O)C(O)C(O)C([O-])=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as c-glycosyl compounds. These are glycoside in which a sugar group is bonded through one carbon to another group via a C-glycosidic bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | C-glycosyl compounds |
|---|
| Alternative Parents | |
|---|
| Substituents | - C-glycosyl compound
- Pentose monosaccharide
- Beta-hydroxy acid
- Sugar acid
- Hydroxy acid
- Monosaccharide
- Tetrahydrofuran
- Secondary alcohol
- Hemiacetal
- Polyol
- Oxacycle
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organoheterocyclic compound
- Primary alcohol
- Alcohol
- Carbonyl group
- Hydrocarbon derivative
- Organic oxide
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | hexuronide and hexuronate degradation | PW000834 | 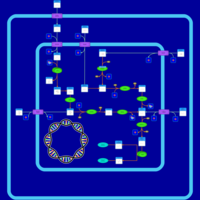 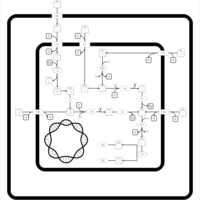 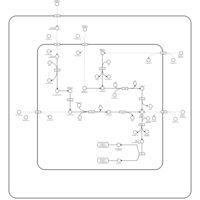 |
|
|---|
| KEGG Pathways: | - Metabolic pathways eco01100
- Pentose and glucuronate interconversions ec00040
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | 439264 | | Kegg ID | C00558 | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|