| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2015-06-04 15:29:43 -0600 |
|---|
| Update Date | 2015-07-23 12:07:15 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Pimeloyl-[acyl-carrier protein] |
|---|
| Description | |
|---|
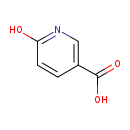
| Structure | |
|---|
| Synonyms: | - 1,6-dihydro-6-oxo-3-Pyridinecarboxylate
- 1,6-dihydro-6-oxo-3-Pyridinecarboxylic acid
- 1,6-dihydro-6-oxo-Nicotinate
- 1,6-dihydro-6-oxo-Nicotinic acid
- 2-Hydroxy-5-carboxypyridine
- 2-Hydroxypyridine-5-carboxylate
- 2-Hydroxypyridine-5-carboxylic acid
- 2-Pyridone-5-carboxylate
- 2-Pyridone-5-carboxylic acid
- 5-Carboxy-2-pyridone
- 6-Hydroxy-nicotinate
- 6-Hydroxy-nicotinic acid
- 6-Hydroxyniacin
- 6-Hydroxynicotinate
- 6-Hydroxynicotinic acid
- 6-Hydroxypyridine-3-carboxylate
- 6-Hydroxypyridine-3-carboxylic acid
- 6-oxo-1H-Pyridine-3-carboxylate
- 6-oxo-1H-Pyridine-3-carboxylic acid
|
|---|
| Chemical Formula: | C6H5NO3 |
|---|
| Weight: | Average: 139.1088
Monoisotopic: 139.026943031 |
|---|
| InChI Key: | BLHCMGRVFXRYRN-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C6H5NO3/c8-5-2-1-4(3-7-5)6(9)10/h1-3H,(H,7,8)(H,9,10) |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | 6-hydroxypyridine-3-carboxylic acid |
|---|
| Traditional IUPAC Name: | 6-hydroxynicotinic acid |
|---|
| SMILES: | OC(=O)C1=CN=C(O)C=C1 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyridinecarboxylic acids. Pyridinecarboxylic acids are compounds containing a pyridine ring bearing a carboxylic acid group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pyridines and derivatives |
|---|
| Sub Class | Pyridinecarboxylic acids and derivatives |
|---|
| Direct Parent | Pyridinecarboxylic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyridine carboxylic acid
- Hydroxypyridine
- Heteroaromatic compound
- Azacycle
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | - Biotin metabolism ec00780
- Drug metabolism - other enzymes ec00983
- Metabolic pathways eco01100
- Microbial metabolism in diverse environments ec01120
- Tropane, piperidine and pyridine alkaloid biosynthesis ec00960
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-014i-1690000000-133fa3cce9e59e8b53a9 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (2 TMS) | splash10-014i-2790000000-e603f3ef71d861b6c58f | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-014i-0290000000-8fed092a7657e583c5d7 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-014i-1690000000-133fa3cce9e59e8b53a9 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-014i-2790000000-e603f3ef71d861b6c58f | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-014i-2790000000-3e2e99cadebe36280a89 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-007a-8900000000-ec493835a384bb643581 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-02mi-9760000000-b9c1d6de28d6f1526e6f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-000i-1900000000-c2874566b889f07322c6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-000f-9800000000-79d5e1067ef5dc7a1ac8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-000f-9800000000-64f4c9b54adbcf50e4f2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0006-9400000000-c052843ee5e55f4051a9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-4e7b7d18474bf24300d6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0fb9-9400000000-bca0e5c9a959c7a9be6c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-056r-9300000000-03e6711f1fe2eb0fba23 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-0a4i-9100000000-e0803e84466b5985c3b2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-0a4i-9000000000-b9b83bf3dff7e6c924fa | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-0006-9000000000-0191434ea4b20d4f055b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0006-1900000000-349f5be72008a76679c5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 30V, Positive | splash10-0096-9600000000-1b3d37aa49ae31d7e29c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0006-9100000000-a528aab22cb1f9af4380 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-1900000000-c2874566b889f07322c6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000f-9800000000-deb0d54a79b61d71a9fc | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000f-9800000000-64f4c9b54adbcf50e4f2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-9400000000-c052843ee5e55f4051a9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-9000000000-4e7b7d18474bf24300d6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0006-9100000000-a528aab22cb1f9af4380 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-006x-0900000000-c48a238051ddd4bb8409 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dl-2900000000-4808d9beefef60fc7a30 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-02fx-9100000000-067bfb3f0aa479a4996c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000l-5900000000-2065745b7d041addf9b2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9400000000-9a8d921204197689240a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-618a7376f25ea052e158 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|