| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-10-10 12:20:18 -0600 |
|---|
| Update Date | 2015-06-03 17:26:04 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | (6S)-6-beta-hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide |
|---|
| Description | (6S)-6-beta-hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide is a substrate for the enzymes ATP-dependent NAD(P)H-hydrate dehydratase which catalyzes the reaction ATP + (6S)-6beta-hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide = ADP + phosphate + NADH. This is an intermediate in NADH "repair". |
|---|
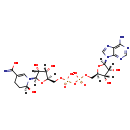
| Structure | |
|---|
| Synonyms: | - (2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-ylmethoxy-hydroxyphosphoryl (2R,3S,4R,5R)-5-(2S)-5-carbamoyl-2-hydroxy-3,4-dihydro-2H-pyridin-1-yl-3,4-dihydroxyoxolan-2-ylmethyl hydrogen phosphate
- (2R,3S,4R,5R)-5-(6-Aminopurin-9-yl)-3,4-dihydroxyoxolan-2-ylmethoxy-hydroxyphosphoryl (2R,3S,4R,5R)-5-(2S)-5-carbamoyl-2-hydroxy-3,4-dihydro-2H-pyridin-1-yl-3,4-dihydroxyoxolan-2-ylmethyl hydrogen phosphoric acid
- (6S)-6-b-Hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide
- (6S)-6-Hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide
- (6S)-6-β-Hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide
- (6S)-6b-Hydroxy-1,4,5,6-tetrahydronicotinamide adenine dinucleotide
- (6S)-6beta-hydroxy-1,4,5,6-tetrahydronicotinamide adenine dinucleotide
- (6S)-6β-Hydroxy-1,4,5,6-tetrahydronicotinamide adenine dinucleotide
- (S)-NADHX
- 6b-Hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide
- 6beta-Hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide
- 6β-Hydroxy-1,4,5,6-tetrahydronicotinamide-adenine dinucleotide
- b-6-HYDROXY-1,4,5,6-tetrhydronicotinamide adenine dinucleotide
- BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE
- β-6-HYDROXY-1,4,5,6-tetrhydronicotinamide adenine dinucleotide
|
|---|
| Chemical Formula: | C21H31N7O15P2 |
|---|
| Weight: | Average: 683.4563
Monoisotopic: 683.135336381 |
|---|
| InChI Key: | IDBZKGQRLBFUFQ-VPHRTNKSSA-N |
|---|
| InChI: | InChI=1S/C21H31N7O15P2/c22-17-12-19(25-6-24-17)28(7-26-12)21-16(33)14(31)10(42-21)5-40-45(37,38)43-44(35,36)39-4-9-13(30)15(32)20(41-9)27-3-8(18(23)34)1-2-11(27)29/h3,6-7,9-11,13-16,20-21,29-33H,1-2,4-5H2,(H2,23,34)(H,35,36)(H,37,38)(H2,22,24,25)/t9-,10-,11+,13-,14-,15-,16-,20-,21-/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | [({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3S,4R,5R)-5-[(2S)-5-carbamoyl-2-hydroxy-1,2,3,4-tetrahydropyridin-1-yl]-3,4-dihydroxyoxolan-2-yl]methoxy})phosphinic acid |
|---|
| Traditional IUPAC Name: | (S)-nadhx |
|---|
| SMILES: | [H][C@]1(COP(O)(=O)OP(O)(=O)OC[C@@]2([H])O[C@@]([H])(N3C=C(CC[C@]3([H])O)C(O)=N)[C@]([H])(O)[C@]2([H])O)O[C@@]([H])(N2C=NC3=C(N)N=CN=C23)[C@]([H])(O)[C@]1([H])O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine nucleotide sugars |
|---|
| Direct Parent | Purine nucleotide sugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine nucleotide sugar
- Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Nicotinamide-nucleotide
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Monoalkyl phosphate
- Tetrahydropyridine
- Imidolactam
- Alkyl phosphate
- Hydropyridine
- Pyrimidine
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Azole
- Vinylogous amide
- Tetrahydrofuran
- Imidazole
- Heteroaromatic compound
- Primary carboxylic acid amide
- Carboxamide group
- Secondary alcohol
- Amino acid or derivatives
- Organoheterocyclic compound
- Oxacycle
- Carboxylic acid derivative
- Azacycle
- Alkanolamine
- Organonitrogen compound
- Organooxygen compound
- Organic nitrogen compound
- Carbonyl group
- Organic oxygen compound
- Organopnictogen compound
- Alcohol
- Primary amine
- Amine
- Organic oxide
- Hydrocarbon derivative
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-017m-1211913000-25814eadfd52e71a50a2 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0920104000-1528abc2c0a942f49031 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900000000-d2229364feb237809db1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1900000000-c059fcd5287ebb902662 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0900316000-b56bcca46a39880ba531 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-1900100000-fd7ebe873ef8a790e710 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a7l-3900000000-e65fb28b52244d9bf64b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00lu-0700009000-84c79a506933cd12dcfe | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00lr-0613159000-d52e45cdb114fde72b48 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0922100000-1592b6a3cfff1659a174 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03e9-0000009000-4a10771d72bd52ae30c5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-057i-6000069000-2683c91f3d01d3ae1e5c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0bvi-6915704000-3bb96fcd514c7f9b7fa8 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | 44236 | | HMDB ID | HMDB0059644 | | Pubchem Compound ID | 440516 | | Kegg ID | C04856 | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available | | Ligand Expo | NAX |
|
|---|