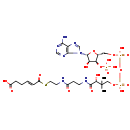| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-10-10 12:19:55 -0600 |
|---|
| Update Date | 2015-06-03 17:26:04 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | 2,3-dehydroadipyl-CoA |
|---|
| Description | 2,3-Dehydroadipyl-CoA is an intermediate in phenylacetate degradation. It is a substrate for the enzyme 2,3-dehydroadipyl-CoA hydratase which catalyzes the reversible conversion of enzymatically produced 2,3-dehydroadipyl-CoA into 3-hydroxyadipyl-CoA. |
|---|
| Structure | |
|---|
| Synonyms: | - (9R,20E)-1-(2R,3S,4R,5R)-5-(6-amino-9H-Purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydro-2-furanyl-3,5,9-trihydroxy-8,8-dimethyl-10,14,19-trioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatetracos
-20-en-24-Oate 3,5-dioxide (non-preferred name)
- (9R,20E)-1-(2R,3S,4R,5R)-5-(6-Amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydro-2-furanyl-3,5,9-trihydroxy-8,8-dimethyl-10,14,19-trioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatetracos
-20-en-24-oic acid 3,5-dioxide (non-preferred name)
- (E)-3,4-dehydroadipoyl-CoA
- (e)-6-2-3-(2R)-4-(2R,3S,4R,5R)-5-(6-Aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-ylmethoxy-hydroxyphosphoryloxy-hydroxyphosphoryloxy-2-hydroxy-3,3-dimethylbutanoylaminopropanoylaminoethylsulfanyl-6-oxohex-4-enoate
- (E)-6-2-3-(2R)-4-(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-ylmethoxy-hydroxyphosphoryloxy-hydroxyphosphoryloxy-2-hydroxy-3,3-dimethylbutanoylaminopropanoylaminoethylsulfanyl-6-oxohex-4-enoic acid
- (e)-6-2-3-(2R)-4-(2R,3S,4R,5R)-5-(6-Aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-ylmethoxy-hydroxyphosphoryloxy-hydroxyphosphoryloxy-2-hydroxy-3,3-dimethylbutanoylaminopropanoylaminoethylsulphanyl-6-oxohex-4-enoate
- (e)-6-2-3-(2R)-4-(2R,3S,4R,5R)-5-(6-Aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-ylmethoxy-hydroxyphosphoryloxy-hydroxyphosphoryloxy-2-hydroxy-3,3-dimethylbutanoylaminopropanoylaminoethylsulphanyl-6-oxohex-4-enoic acid
- 2,3-Dehydroadipyl-CoA
- 2,3-Didehydroadipyl-CoA
- 5-Carboxy-2-pentenoyl-CoA
|
|---|
| Chemical Formula: | C27H42N7O19P3S |
|---|
| Weight: | Average: 893.644
Monoisotopic: 893.146902423 |
|---|
| InChI Key: | ZFXICKRXPZTFPB-KCQRSJHASA-N |
|---|
| InChI: | InChI=1S/C27H42N7O19P3S/c1-27(2,22(40)25(41)30-8-7-16(35)29-9-10-57-18(38)6-4-3-5-17(36)37)12-50-56(47,48)53-55(45,46)49-11-15-21(52-54(42,43)44)20(39)26(51-15)34-14-33-19-23(28)31-13-32-24(19)34/h4,6,13-15,20-22,26,39-40H,3,5,7-12H2,1-2H3,(H,29,35)(H,30,41)(H,36,37)(H,45,46)(H,47,48)(H2,28,31,32)(H2,42,43,44)/b6-4+/t15-,20-,21-,22+,26-/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (4E)-6-[(2-{3-[(2R)-3-[({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-2-hydroxy-3-methylbutanamido]propanamido}ethyl)sulfanyl]-6-oxohex-4-enoic acid |
|---|
| Traditional IUPAC Name: | trans-2,3-didehydroadipoyl-coa |
|---|
| SMILES: | O[C@H](C(C)(C)COP(=O)(O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N)C(=O)NCCC(=O)NCCSC(=O)\C=C\CCC(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as aromatic monoterpenoids. These are monoterpenoids containing at least one aromatic ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Monoterpenoids |
|---|
| Direct Parent | Aromatic monoterpenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Aromatic monoterpenoid
- Hydroxybenzoic acid
- Monocyclic monoterpenoid
- Benzoic acid or derivatives
- Benzoic acid
- Benzoyl
- 1-hydroxy-2-unsubstituted benzenoid
- Phenol
- Monocyclic benzene moiety
- Benzenoid
- Carboxylic acid
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Organic oxygen compound
- Hydrocarbon derivative
- Organic oxide
- Organooxygen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -5 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1922000130-7f64797205f4c5a0799d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0933000000-02ff8b4f16369cd99c6a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-2911000000-dccaccec5a4ef0c29d6e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0059-3911030450-c20f26f9727f01bfff34 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-003r-3911010010-c72024d64ac9c2d76bb0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-057i-6900100000-c0c56fb21cc884106020 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004u-0200000090-be9fb26b71e54a88c466 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0901000160-224040439203b998045f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0119000000-17a09959c8b36d05b052 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0000000090-5ff647d6f62bf4c645f4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0597-5800001690-98c1caedf251d7207161 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004l-9202423730-7094fc194ad3dbe28e06 | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Yurtsever D. (2007). Fatty acid methyl ester profiling of Enterococcus and Esherichia coli for microbial source tracking. M.sc. Thesis. Villanova University: U.S.A
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|