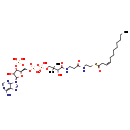| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-10-10 12:19:06 -0600 |
|---|
| Update Date | 2015-06-03 17:26:02 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | (3Z)-Dodec-3-enoyl-CoA |
|---|
| Description | 3Z-Dodecenoyl-CoA is an intermediate in fatty acid metabolism. 3Z-dodecenoyl-CoA is converted from trans-Dodec-2-enoyl-CoA via acyl-CoA oxidase, acyl-CoA dehydrogenase, and long-chain-acyl-CoA dehydrogenase. |
|---|
| Structure | |
|---|
| Synonyms: | - (3Z)-dodec-3-enoyl-CoA
- (3Z)-dodec-3-enoyl-Coenzyme A
- (3Z)-dodecenoyl-CoA
- 3-cis-Dodecenoyl coenzyme A
- 3-cis-Dodecenoyl coenzyme A
- S-[2-[3-[[(2R)-4-[[[(2R,3R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-hydroxyphosphoryl]oxy-2-hydroxy-3,3-dimethylbutanoyl]amino]propanoylamino]ethyl] (Z)-dodec-3-enethioate
- S-[2-[3-[[(2R)-4-[[[(2R,3R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-phosphonooxyoxolan-2-yl]methoxy-hydroxyphosphoryl]oxy-hydroxyphosphoryl]oxy-2-hydroxy-3,3-dimethylbutanoyl]amino]propanoylamino]ethyl] (Z)-dodec-3-enethioic acid
|
|---|
| Chemical Formula: | C33H56N7O17P3S |
|---|
| Weight: | Average: 947.821
Monoisotopic: 947.266623627 |
|---|
| InChI Key: | XEMIVMKTVGRFTD-BFJLDWRUSA-N |
|---|
| InChI: | InChI=1S/C33H56N7O17P3S/c1-4-5-6-7-8-9-10-11-12-13-24(42)61-17-16-35-23(41)14-15-36-31(45)28(44)33(2,3)19-54-60(51,52)57-59(49,50)53-18-22-27(56-58(46,47)48)26(43)32(55-22)40-21-39-25-29(34)37-20-38-30(25)40/h11-12,20-22,26-28,32,43-44H,4-10,13-19H2,1-3H3,(H,35,41)(H,36,45)(H,49,50)(H,51,52)(H2,34,37,38)(H2,46,47,48)/b12-11-/t22-,26?,27+,28+,32-/m1/s1 |
|---|
| CAS number: | 2152-91-2 |
|---|
| IUPAC Name: | {[(2R,3R,5R)-5-(6-amino-9H-purin-9-yl)-2-({[({[(3R)-3-{[2-({2-[(3Z)-dodec-3-enoylsulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}-3-hydroxy-2,2-dimethylpropoxy](hydroxy)phosphoryl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-hydroxyoxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional IUPAC Name: | (3Z)-dodecenoyl-coa |
|---|
| SMILES: | CCCCCCCC\C=C/CC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H](C(O)[C@H]1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 3-enoyl coas. These are organic compounds containing a coenzyme A substructure linked to a 3-enoyl chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | 3-enoyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -4 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1911101102-f755a7227f8f5cf26a51 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0931300000-e8ee2e6ba89c440c4b01 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1900001000-1e343c2de4ba69b0ba33 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0059-3922031304-940eef4ed15c69c25b44 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0059-2901110001-884e4dded5bd961752aa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-057i-6900100000-e276101429e1dde72045 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0000000009-e0195a9abcfc5c3ca328 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-003r-2000201679-a1b7c61a017c1da9109e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-0002900000-da544c65a54f9d98d6e5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0000000009-983824b3dc4935215fda | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-4200302409-b3dbdc37620a95871ac3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00p0-4203502906-cc5ba47b21cb235b235d | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Yurtsever D. (2007). Fatty acid methyl ester profiling of Enterococcus and Esherichia coli for microbial source tracking. M.sc. Thesis. Villanova University: U.S.A
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|