| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-10-10 12:17:39 -0600 |
|---|
| Update Date | 2015-06-03 17:25:58 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
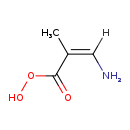
| Name: | (Z)-2-Methyl-peroxyaminoacrylate |
|---|
| Description | (Z)-2-Methyl-peroxyaminoacrylate is an intermediate in the degradation of exogenous pyrimidines via the RutB pathway. It is a substrate for the enzyme Peroxyureidoacrylate/ureidoacrylate amidohydrolase which catalyzes the reaction (Z)-3-ureidoacrylate peracid + H(2)O = (Z)-3-peroxyaminoacrylate + NH(3). This enzyme quickly hydrolyzes the ureidoacrylate peracid to avoid toxicity, but can also hydrolyzes ureidoacrylate that is formed spontaneously from ureidoacrylate peracid. |
|---|
| Structure | |
|---|
| Synonyms: | - (2Z)-3-amino-2-Methyl-2-propeneperoxoate
- (2Z)-3-Amino-2-methyl-2-propeneperoxoic acid
- (Z)-2-hydroxy-3-peroxyaminoacrylate
- (Z)-2-Hydroxy-3-peroxyaminoacrylic acid
- (Z)-2-Methyl-peroxyaminoacrylic acid
- (Z)-2-Methylperoxyaminoacrylate
- (Z)-2-Methylperoxyaminoacrylic acid
- (Z)-3-amino-2-Methylprop-2-eneperoxoate
- (Z)-3-amino-2-methylprop-2-eneperoxoic acid
|
|---|
| Chemical Formula: | C4H7NO3 |
|---|
| Weight: | Average: 117.1033
Monoisotopic: 117.042593095 |
|---|
| InChI Key: | DYYSAMFOJRGAMQ-IHWYPQMZSA-N |
|---|
| InChI: | InChI=1S/C4H7NO3/c1-3(2-5)4(6)8-7/h2,7H,5H2,1H3/b3-2- |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2Z)-3-amino-2-methylprop-2-eneperoxoic acid |
|---|
| Traditional IUPAC Name: | (2Z)-3-amino-2-methylprop-2-eneperoxoic acid |
|---|
| SMILES: | [H]\C(N)=C(/C)C(=O)OO |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as peroxycarboxylic acids. These are organic acids with the general formula [H]OOC(R)=O (R = H, organyl group). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Peroxycarboxylic acids and derivatives |
|---|
| Direct Parent | Peroxycarboxylic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Peroxycarboxylic acid
- Vinylogous amide
- Amino acid or derivatives
- Carboxylic acid salt
- Hydroperoxide
- Enamine
- Monocarboxylic acid or derivatives
- Peroxol
- Allylamine
- Organic oxide
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Amine
- Primary aliphatic amine
- Organic oxygen compound
- Primary amine
- Organic salt
- Carbonyl group
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 0 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-067i-9800000000-85c460292b4daaabdd31 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05o0-9200000000-b5da1624185ddca33d03 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-9000000000-d7bcd160e640a4faf27a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-3900000000-aa48dc8acc25f5651923 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0159-9500000000-c1cce8096eb976a6f11b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0uyi-9000000000-96cc642ea5aae042e2d1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9100000000-ff794d5b0dee2ae71c18 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9000000000-e72e92c1dbcd2e48e59d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-9000000000-2b671d7ac52b830cbafd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9200000000-42f0a2ebd8b75ef48eaa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001j-9100000000-04957a91bf9579c747c9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0zfr-9000000000-ee4f7adb0930dde995d1 | View in MoNA |
|---|
|
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|