| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-10-10 12:12:11 -0600 |
|---|
| Update Date | 2015-06-03 17:25:43 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
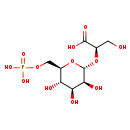
| Name: | 2-O-(6-Phospho-alpha-D-mannosyl)-D-glycerate |
|---|
| Description | 2-O-(6-phospho-alpha-D-mannosyl)-D-glycerate is a substrate for 2-O-(6-phospho-alpha-D-mannosyl)-D-glycerate acylhydrolase. The enzyme participates in the mannosylglycerate degradation pathway of some bacteria. Mannosylglycerate is phosphorylated during transport into the cell, and the phosphorylated form is hydrolysed by this enzyme. It is involved in 2-O-alpha-mannosyl-D-glycerate degradation. |
|---|
| Structure | |
|---|
| Synonyms: | - (2R)-3-Hydroxy-2-(2R,3S,4S,5S,6R)-3,4,5-trihydroxy-6-(phosphonooxymethyl)oxan-2-yloxypropanoate
- (2R)-3-hydroxy-2-(2R,3S,4S,5S,6R)-3,4,5-trihydroxy-6-(phosphonooxymethyl)oxan-2-yloxypropanoic acid
- (2R)-3-Hydroxy-2-(6-O-phosphono-a-D-mannopyranosyl)oxypropanoate
- (2R)-3-Hydroxy-2-(6-O-phosphono-a-D-mannopyranosyl)oxypropanoic acid
- (2R)-3-Hydroxy-2-(6-O-phosphono-alpha-D-mannopyranosyl)oxypropanoate
- (2R)-3-Hydroxy-2-(6-O-phosphono-alpha-D-mannopyranosyl)oxypropanoic acid
- (2R)-3-Hydroxy-2-(6-O-phosphono-α-D-mannopyranosyl)oxypropanoate
- (2R)-3-Hydroxy-2-(6-O-phosphono-α-D-mannopyranosyl)oxypropanoic acid
- 2(α-D-mannosyl-6-phosphate)-D-glycerate
- 2(α-D-mannosyl-6-phosphoric acid)-D-glyceric acid
- 2-O-(6-phospho-a-D-Mannosyl)-D-glycerate
- 2-O-(6-phospho-a-D-Mannosyl)-D-glyceric acid
- 2-O-(6-Phospho-a-mannosyl)-D-glycerate
- 2-O-(6-Phospho-a-mannosyl)-D-glyceric acid
- 2-O-(6-phospho-a-Mannosyl)-delta-glycerate
- 2-O-(6-phospho-a-Mannosyl)-delta-glyceric acid
- 2-O-(6-phospho-a-Mannosyl)-δ-glycerate
- 2-O-(6-phospho-a-Mannosyl)-δ-glyceric acid
- 2-O-(6-phospho-alpha-D-Mannosyl)-D-glyceric acid
- 2-O-(6-Phospho-alpha-mannosyl)-D-glycerate
- 2-O-(6-Phospho-alpha-mannosyl)-D-glyceric acid
- 2-O-(6-Phospho-alpha-mannosyl)-delta-glycerate
- 2-O-(6-Phospho-alpha-mannosyl)-delta-glyceric acid
- 2-O-(6-phospho-α-D-Mannosyl)-D-glycerate
- 2-O-(6-phospho-α-D-Mannosyl)-D-glyceric acid
- 2-O-(6-phospho-α-Mannosyl)-D-glycerate
- 2-O-(6-phospho-α-Mannosyl)-D-glyceric acid
- 2-O-(6-phospho-α-Mannosyl)-δ-glycerate
- 2-O-(6-phospho-α-Mannosyl)-δ-glyceric acid
|
|---|
| Chemical Formula: | C9H17O12P |
|---|
| Weight: | Average: 348.1978
Monoisotopic: 348.04576252 |
|---|
| InChI Key: | BOLXAGHGKNGVBE-MTXRGOKVSA-N |
|---|
| InChI: | InChI=1S/C9H17O12P/c10-1-3(8(14)15)20-9-7(13)6(12)5(11)4(21-9)2-19-22(16,17)18/h3-7,9-13H,1-2H2,(H,14,15)(H2,16,17,18)/t3-,4-,5-,6+,7+,9+/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2R)-3-hydroxy-2-{[(2R,3S,4S,5S,6R)-3,4,5-trihydroxy-6-[(phosphonooxy)methyl]oxan-2-yl]oxy}propanoic acid |
|---|
| Traditional IUPAC Name: | (2R)-3-hydroxy-2-{[(2R,3S,4S,5S,6R)-3,4,5-trihydroxy-6-[(phosphonooxy)methyl]oxan-2-yl]oxy}propanoic acid |
|---|
| SMILES: | OC[C@@H](O[C@H]1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H](O)[C@@H]1O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose phosphate
- Fatty acyl glycoside
- Fatty acyl glycoside of mono- or disaccharide
- Glycosyl compound
- O-glycosyl compound
- Monosaccharide phosphate
- Beta-hydroxy acid
- Monoalkyl phosphate
- Glyceric_acid
- Sugar acid
- Alkyl phosphate
- Fatty acyl
- Hydroxy acid
- Organic phosphoric acid derivative
- Oxane
- Phosphoric acid ester
- Secondary alcohol
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Acetal
- Oxacycle
- Carboxylic acid
- Carboxylic acid derivative
- Polyol
- Hydrocarbon derivative
- Organic oxide
- Alcohol
- Carbonyl group
- Primary alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -3 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | 2-O-alpha-mannosyl-D-glycerate degradation | PW002096 |    |
|
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03ea-9624000000-b2eb9ad809dfb1614adc | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-00di-3530239000-c31601470a21a35a479d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_4_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_4_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("2-O-(6-Phospho-alpha-mannosyl)-D-glycerate,3TBDMS,#8" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-053j-9437000000-c5fa5dddffe828fd1b0e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-052r-9310000000-ada2af57904347782db9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4r-9600000000-d6cbf6642be6233baf9b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-054t-9324000000-66e5bfc65619e1fbc660 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9100000000-fff4af92a3b80c0f63c9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-4927cd7fdf9692a12431 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-1391000000-9b1fea4aa36b2edf8641 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01pa-9620000000-e994296d32e32ffdaeb4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05fs-9300000000-c68a692330ed21620ab7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002b-9011000000-4784bc2eb78346de6c49 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-9010000000-ea3e1896d17fa5092d85 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-849c1c60f1e41f399334 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Ryu J, Kanapathipillai M, Lentzen G, Park CB: Inhibition of beta-amyloid peptide aggregation and neurotoxicity by alpha-d-mannosylglycerate, a natural extremolyte. Peptides. 2008 Apr;29(4):578-84. Epub 2008 Jan 9. Pubmed: 18304694
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|