| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-08-09 09:25:09 -0600 |
|---|
| Update Date | 2015-06-03 17:21:39 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
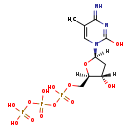
| Name: | 5-Methyldeoxycytidine triphosphate |
|---|
| Description | 5-methyldeoxycytidine triphosphate is a member of the chemical class known as Pyrimidine 2'-deoxyribonucleoside Triphosphates. These are pyrimidine nucleotides with a triphosphate group linked to the ribose moiety lacking an hydroxyl group at position 2. This is a methylated form of dCTP. Methylation of nucleotides and nucleosides can occur spontaneously through the action of alkylating reagents. |
|---|
| Structure | |
|---|
| Synonyms: | - 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE)
- 2'-DEOXY-5-methylcytidine 5'-(tetrahydrogen triphosphoric acid)
- 5-Methyl deoxycytidine-5'-triphosphate
- 5-Methyl deoxycytidine-5'-triphosphoric acid
- 5-Methyl deoxycytidine-triphosphate
- 5-Methyl deoxycytidine-triphosphoric acid
- 5-Methyl-2'-deoxycytidine 5'-triphosphate
- 5-Methyl-2'-deoxycytidine 5'-triphosphoric acid
- 5-Methyl-dCTP
- 5-Methyldeoxycytidine triphosphate
- 5-Methyldeoxycytidine triphosphoric acid
|
|---|
| Chemical Formula: | C10H18N3O13P3 |
|---|
| Weight: | Average: 481.1835
Monoisotopic: 481.005247213 |
|---|
| InChI Key: | NGYHUCPPLJOZIX-XLPZGREQSA-N |
|---|
| InChI: | InChI=1S/C10H18N3O13P3/c1-5-3-13(10(15)12-9(5)11)8-2-6(14)7(24-8)4-23-28(19,20)26-29(21,22)25-27(16,17)18/h3,6-8,14H,2,4H2,1H3,(H,19,20)(H,21,22)(H2,11,12,15)(H2,16,17,18)/t6-,7+,8+/m0/s1 |
|---|
| CAS number: | 22003-12-9 |
|---|
| IUPAC Name: | {[hydroxy({[hydroxy({[(2R,3S,5R)-3-hydroxy-5-(2-hydroxy-4-imino-5-methyl-1,4-dihydropyrimidin-1-yl)oxolan-2-yl]methoxy})phosphoryl]oxy})phosphoryl]oxy}phosphonic acid |
|---|
| Traditional IUPAC Name: | [hydroxy({hydroxy[(2R,3S,5R)-3-hydroxy-5-(2-hydroxy-4-imino-5-methylpyrimidin-1-yl)oxolan-2-yl]methoxyphosphoryl}oxy)phosphoryl]oxyphosphonic acid |
|---|
| SMILES: | [H][C@]1(O)C[C@@]([H])(O[C@]1([H])COP(O)(=O)OP(O)(=O)OP(O)(O)=O)N1C=C(C)C(=N)N=C1O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine 2'-deoxyribonucleoside triphosphates. These are pyrimidine nucleotides with a triphosphate group linked to the ribose moiety lacking a hydroxyl group at position 2. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine deoxyribonucleotides |
|---|
| Direct Parent | Pyrimidine 2'-deoxyribonucleoside triphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine 2'-deoxyribonucleoside triphosphate
- Aminopyrimidine
- Pyrimidone
- Monoalkyl phosphate
- Hydropyrimidine
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Pyrimidine
- Imidolactam
- Alkyl phosphate
- Heteroaromatic compound
- Tetrahydrofuran
- Secondary alcohol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Primary amine
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Alcohol
- Amine
- Organic nitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -3 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | 61766 | | HMDB ID | Not Available | | Pubchem Compound ID | 161376 | | Kegg ID | Not Available | | ChemSpider ID | 141756 | | Wikipedia ID | Not Available | | BioCyc ID | Not Available | | Ligand Expo | 523 |
|
|---|