Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
7-Aminomethyl-7-carbaguanine (M2MDB001851)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-08-09 09:16:18 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:21:33 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | 7-Aminomethyl-7-carbaguanine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | 7-Aminomethyl-7-carbaguanine is one of the precursors of nucleoside Q (queuosine) biosynthesis. It is a substrate for preQ1 synthase (EC 1.7.1.13) which catalyzes the NADPH-dependent reduction of 7-cyano-7-carbaguanine (preQ0) to 7-aminomethyl-7-carbaguanine (preQ1). More specifically, this enzyme catalyzes the chemical reaction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
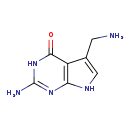
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C7H9N5O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 179.1793 Monoisotopic: 179.080709935 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | MEYMBLGOKYDGLZ-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C7H9N5O/c8-1-3-2-10-5-4(3)6(13)12-7(9)11-5/h2H,1,8H2,(H4,9,10,11,12,13) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2-amino-5-(aminomethyl)-3H,4H,7H-pyrrolo[2,3-d]pyrimidin-4-one | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 7-aminomethyl-7-deazaguanine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | NCC1=CNC2=C1C(=O)NC(N)=N2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as pyrrolo[2,3-d]pyrimidines. These are aromatic heteropolycyclic compounds containing a pyrrolo[2,3-d]pyrimidine ring system, which is an pyrrolopyrimidine isomers having the 3 ring nitrogen atoms at the 1-, 5-, and 7-positions. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Pyrrolopyrimidines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Pyrrolo[2,3-d]pyrimidines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pyrrolo[2,3-d]pyrimidines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 7-Aminomethyl-7-carbaguanine + 2 NADP > 7-Cyano-7-carbaguanine +2 NADPH Guanine(34) in tRNA + 7-Aminomethyl-7-carbaguanine > 7-aminomethyl-7-carbaguanine(34) in tRNA + Guanine Queuine + 7-Aminomethyl-7-carbaguanine <> Guanine 7-Aminomethyl-7-carbaguanine + NADP <> 7-Cyano-7-carbaguanine + NADPH +2 Hydrogen ion 7-Cyano-7-carbaguanine + 3 Hydrogen ion + 2 NADPH + 2 NADPH >2 NADP + 7-Aminomethyl-7-carbaguanine 7-Aminomethyl-7-carbaguanine + tRNA guanine > 7-aminomethyl-7-deazaguanosine34 in tRNA + Guanine 7 7-Aminomethyl-7-carbaguanine + NADP <>7 7-Cyano-7-carbaguanine + NADPH +2 Hydrogen ion Queuine + 7 7-Aminomethyl-7-carbaguanine <> Guanine Queuine + 7 7-Aminomethyl-7-carbaguanine <> Guanine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in queuine tRNA-ribosyltransferase activity
- Specific function:
- Exchanges the guanine residue with 7-aminomethyl-7- deazaguanine in tRNAs with GU(N) anticodons (tRNA-Asp, -Asn, -His and -Tyr). After this exchange, a cyclopentendiol moiety is attached to the 7-aminomethyl group of 7-deazaguanine, resulting in the hypermodified nucleoside queuosine (Q) (7-(((4,5-cis- dihydroxy-2-cyclopenten-1-yl)amino)methyl)-7-deazaguanosine)
- Gene Name:
- tgt
- Uniprot ID:
- P0A847
- Molecular weight:
- 42593
Reactions
| [tRNA]-guanine + queuine = [tRNA]-queuine + guanine. |
| [tRNA]-guanine + 7-aminomethyl-7-carbaguanine = [tRNA]-7-aminomethyl-7-carbaguanine + guanine. |
- General function:
- Involved in oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor
- Specific function:
- Catalyzes the NADPH-dependent reduction of 7-cyano-7- deazaguanine (preQ0) to 7-aminomethyl-7-deazaguanine (preQ1)
- Gene Name:
- queF
- Uniprot ID:
- Q46920
- Molecular weight:
- 32587
Reactions
| 7-aminomethyl-7-carbaguanine + 2 NADP(+) = 7-cyano-7-carbaguanine + 2 NADPH. |