| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-08-09 09:16:13 -0600 |
|---|
| Update Date | 2015-06-03 17:21:29 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Diadenosine triphosphate |
|---|
| Description | Diadenosine triphosphate (AP3A) is a species of diadenosine polyphosphate (ApnA), consisting of two adenosines joined by a chain of phosphates. (inferred from compound structure) In E. coli, AP3A can be produced from ATP and ADP in a reaction catalyzed by AP3A synthetase, which is encoded by the gene lysU. (EcoCyc). These dinucleotides have been proposed to act as modulators of the heat-shock response and stress response. |
|---|
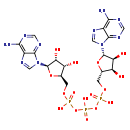
| Structure | |
|---|
| Synonyms: | - 5'Ap3A
- Adenosine (5')triphospho(5')adenosine
- Adenosine 5'-(tetrahydrogen triphosphate), P''-5'-ester with adenosine
- Adenosine 5'-(tetrahydrogen triphosphoric acid), p''-5'-ester with adenosine
- Adenosine 5'-triphosphate 5'-adenosine
- Adenosine 5'-triphosphoric acid 5'-adenosine
- Adenosine(3)triphosphate adenosine
- Adenosine(3)triphosphoric acid adenosine
- Adenosine(5')triphospho(5')adenosine
- Ap3a
- Ap3A
- BIS(ADENOSINE)-5'-TRIPHOSPHATE
- BIS(adenosine)-5'-triphosphoric acid
- Diadenosine triphosphate
- Diadenosine triphosphoric acid
- P(1),P(3)-bis(5'-adenosyl) trihydrogen triphosphate
- P(1),P(3)-Bis(5'-adenosyl) trihydrogen triphosphoric acid
- P(1),P(3)-bis(5'-adenosyl) triphosphate
- P(1),P(3)-Bis(5'-adenosyl) triphosphoric acid
- P(1)-P(3)-bis(5'-adenosyl) triphosphate
- P(1)-P(3)-Bis(5'-adenosyl) triphosphoric acid
- P1,P3-Bis(5'-adenosyl) triphosphate
- P1,P3-Bis(5'-adenosyl) triphosphoric acid
- P1,P3-bis(5'-adenosyl)triphosphate
- P1,P3-Bis(5'-adenosyl)triphosphoric acid
|
|---|
| Chemical Formula: | C20H27N10O16P3 |
|---|
| Weight: | Average: 756.4071
Monoisotopic: 756.081934402 |
|---|
| InChI Key: | QCICUPZZLIQAPA-WGIMJHEJSA-N |
|---|
| InChI: | InChI=1S/C20H27N10O16P3/c21-15-9-17(25-3-23-15)29(5-27-9)19-13(33)11(31)7(43-19)1-41-47(35,36)45-49(39,40)46-48(37,38)42-2-8-12(32)14(34)20(44-8)30-6-28-10-16(22)24-4-26-18(10)30/h3-8,11-14,19-20,31-34H,1-2H2,(H,35,36)(H,37,38)(H,39,40)(H2,21,23,25)(H2,22,24,26)/t7-,8+,11-,12+,13-,14+,19-,20+ |
|---|
| CAS number: | 56432-02-1 |
|---|
| IUPAC Name: | {[(2S,3R,4S,5S)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)phosphinic acid |
|---|
| Traditional IUPAC Name: | [(2S,3R,4S,5S)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy({[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxyphosphinic acid |
|---|
| SMILES: | NC1=C2N=CN([C@H]3O[C@@H](COP(O)(=O)OP(O)(=O)OP(O)(=O)OC[C@H]4O[C@H]([C@H](O)[C@@H]4O)N4C=NC5=C(N)N=CN=C45)[C@H](O)[C@@H]3O)C2=NC=N1 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as very long-chain fatty acids. These are fatty acids with an aliphatic tail that contains at least 22 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Very long-chain fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Very long-chain fatty acid
- Straight chain fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -3 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0ab9-1211090100-8dd4ac3e3f704ce81480 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0910100300-a4645b9f9e4bff03904a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900000000-9d576618c3405dd82c62 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0900000000-d874a1705ef911a68a3f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a59-0700110900-6003bb661e58b16c00f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-0900100000-cc9f72dc9a076e69680c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001r-1940100000-375a011d350537091bbb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000000900-6de9f94edaaf63b06abb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-1302302900-67b889b6aeb6c6b75d7f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0600901100-01ba1d8c1675b37b46f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0100000900-f690574ac6b3670ec7c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-0300003900-7ccaa7c6ba1c635f1660 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0901010000-452975aa0aa202f669c0 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Baker MD, Holloway DE, Swaminathan GJ, Acharya KR: Crystal structures of eosinophil-derived neurotoxin (EDN) in complex with the inhibitors 5'-ATP, Ap3A, Ap4A, and Ap5A. Biochemistry. 2006 Jan 17;45(2):416-26. Pubmed: 16401072
- Hollah P, Hausberg M, Kosch M, Barenbrock M, Letzel M, Schlatter E, Rahn KH: A novel assay for determination of diadenosine polyphosphates in human platelets: studies in normotensive subjects and in patients with essential hypertension. J Hypertens. 2001 Feb;19(2):237-45. Pubmed: 11212966
- Jankowski J, Jankowski V, Laufer U, van der Giet M, Henning L, Tepel M, Zidek W, Schluter H: Identification and quantification of diadenosine polyphosphate concentrations in human plasma. Arterioscler Thromb Vasc Biol. 2003 Jul 1;23(7):1231-8. Epub 2003 May 8. Pubmed: 12738682
- Kisselev LL, Justesen J, Wolfson AD, Frolova LY: Diadenosine oligophosphates (Ap(n)A), a novel class of signalling molecules? FEBS Lett. 1998 May 8;427(2):157-63. Pubmed: 9607303
- Kowara R, Karaczyn AA, Fivash MJ Jr, Kasprzak KS: In vitro inhibition of the enzymatic activity of tumor suppressor FHIT gene product by carcinogenic transition metals. Chem Res Toxicol. 2002 Mar;15(3):319-25. Pubmed: 11896678
- Pintor J, Carracedo G, Alonso MC, Bautista A, Peral A: Presence of diadenosine polyphosphates in human tears. Pflugers Arch. 2002 Jan;443(3):432-6. Epub 2001 Aug 23. Pubmed: 11810214
- Pintor J, King BF, Miras-Portugal MT, Burnstock G: Selectivity and activity of adenine dinucleotides at recombinant P2X2 and P2Y1 purinoceptors. Br J Pharmacol. 1996 Nov;119(5):1006-12. Pubmed: 8922753
- Turpaev K, Hartmann R, Kisselev L, Justesen J: Ap3A and Ap4A are primers for oligoadenylate synthesis catalyzed by interferon-inducible 2-5A synthetase. FEBS Lett. 1997 May 19;408(2):177-81. Pubmed: 9187362
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Vartanian AA: Gelsolin and plasminogen activator inhibitor-1 are Ap3A-binding proteins. Ital J Biochem. 2003 Mar;52(1):9-16. Pubmed: 12833632
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|