| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-08-03 18:24:20 -0600 |
|---|
| Update Date | 2015-09-17 16:24:39 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Cytidine monophosphate N-acetylneuraminic acid |
|---|
| Description | Cytidine monophosphate N-acetylneuraminic acid is nucleoside monophosphate sugar which donates N-acetylneuraminic acid to the terminal sugar of a glycoprotein. It is an intermediate in the synthesis of peptidoglycan. |
|---|
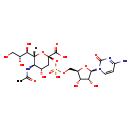
| Structure | |
|---|
| Synonyms: | - CMP-N-acetylneuraminate
- CMP-N-Acetylneuraminic acid
- CMP-N-acylneuraminate
- CMP-N-Acylneuraminic acid
- CMP-NeuNAc
- Cytidine 5'-monophosphate N-acetylneuraminate
- Cytidine 5'-monophosphate N-acetylneuraminic acid
- Cytidine 5'-monophosphoric acid N-acetylneuraminic acid
- Cytidine monophosphate N-acetylneuraminate
- Cytidine monophosphoric acid N-acetylneuraminic acid
|
|---|
| Chemical Formula: | C20H31N4O16P |
|---|
| Weight: | Average: 614.4511
Monoisotopic: 614.147267476 |
|---|
| InChI Key: | TXCIAUNLDRJGJZ-GDTVTMGESA-N |
|---|
| InChI: | InChI=1S/C20H31N4O16P/c1-7(26)22-12-8(27)4-20(18(32)33,39-16(12)13(29)9(28)5-25)40-41(35,36)37-6-10-14(30)15(31)17(38-10)24-3-2-11(21)23-19(24)34/h2-3,8-10,12-17,25,27-31H,4-6H2,1H3,(H,22,26)(H,32,33)(H,35,36)(H2,21,23,34)/t8?,9-,10-,12-,13-,14-,15-,16-,17-,20-/m1/s1 |
|---|
| CAS number: | 22-12-8 |
|---|
| IUPAC Name: | (2R,4S,5R,6R)-2-[({[(2R,3S,4R,5R)-5-(4-amino-2-oxo-1,2-dihydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]-5-acetamido-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]oxane-2-carboxylic acid |
|---|
| Traditional IUPAC Name: | cmp-sialic acid |
|---|
| SMILES: | [H][C@@](O)(CO)[C@@]([H])(O)[C@]1([H])O[C@](C[C@]([H])(O)[C@@]1([H])NC(C)=O)(OP(O)(=O)OC[C@@]1([H])O[C@@]([H])(N2C=CC(N)=NC2=O)[C@]([H])(O)[C@]1([H])O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine nucleotide sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine nucleotide sugars |
|---|
| Direct Parent | Pyrimidine nucleotide sugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine nucleotide sugar
- Pyrimidine ribonucleoside monophosphate
- N-acylneuraminic acid
- N-acylneuraminic acid or derivatives
- Neuraminic acid
- Pentose phosphate
- Pentose-5-phosphate
- C-glucuronide
- C-glycosyl compound
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Aminopyrimidine
- Pyrimidone
- Dialkyl phosphate
- Hydropyrimidine
- Monosaccharide
- Organic phosphoric acid derivative
- Oxane
- Phosphoric acid ester
- Pyran
- Pyrimidine
- Imidolactam
- Alkyl phosphate
- Tetrahydrofuran
- Acetamide
- Heteroaromatic compound
- Carboxamide group
- Secondary carboxylic acid amide
- Secondary alcohol
- Amino acid
- Amino acid or derivatives
- Polyol
- Organoheterocyclic compound
- Azacycle
- Carboxylic acid derivative
- Carboxylic acid
- Oxacycle
- Monocarboxylic acid or derivatives
- Organopnictogen compound
- Organic oxide
- Carbonyl group
- Organic oxygen compound
- Primary amine
- Alcohol
- Primary alcohol
- Hydrocarbon derivative
- Amine
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| | Concentration | Strain | Media | Growth Status | Growth System | Temperature | Details |
|---|
| 169± 0 uM | BW25113 | 48 mM Na2HPO4, 22 mM KH2PO4, 10 mM NaCl, 45 mM (NH4)2SO4, supplemented with 1 mM MgSO4, 1 mg/l thiamine·HCl, 5.6 mg/l CaCl2, 8 mg/l FeCl3, 1 mg/l MnCl2·4H2O, 1.7 mg/l ZnCl2, 0.43 mg/l CuCl2·2H2O, 0.6 mg/l CoCl2·2H2O and 0.6 mg/l Na2MoO4·2H2O. 4 g/L Gluco | Stationary Phase, glucose limited | Bioreactor, pH controlled, O2 and CO2 controlled, dilution rate: 0.2/h | 37 oC | PMID: 17379776 |
|
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01ot-9512160000-178f009287639a11dbfd | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-044l-9624017000-8c596d9b4644a97fbb65 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0912020000-9d330aadc1882c503212 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-3902000000-b2c85490ddb7f53db6f4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-6910000000-a8d1f94c4661e1dd3097 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-4813091000-d68fb03c5bbe9978f2e5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-4912010000-168821fb3a4dc5a309b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-08fr-9310000000-5e2455827ac9cc8b0a87 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-2004169000-eac426191a551fa0ea5f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0bvs-9003180000-afd899893a04da29e2e4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004l-9001310000-a0291ac7ad84d4ba3ecb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0240369000-cae65d161602c1ecc345 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-1864591000-a0ca8cc5e78443e5226c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03k9-3943120000-4ac27539b86275883867 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Ishii, N., Nakahigashi, K., Baba, T., Robert, M., Soga, T., Kanai, A., Hirasawa, T., Naba, M., Hirai, K., Hoque, A., Ho, P. Y., Kakazu, Y., Sugawara, K., Igarashi, S., Harada, S., Masuda, T., Sugiyama, N., Togashi, T., Hasegawa, M., Takai, Y., Yugi, K., Arakawa, K., Iwata, N., Toya, Y., Nakayama, Y., Nishioka, T., Shimizu, K., Mori, H., Tomita, M. (2007). "Multiple high-throughput analyses monitor the response of E. coli to perturbations." Science 316:593-597. Pubmed: 17379776
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | 16556 | | HMDB ID | HMDB01176 | | Pubchem Compound ID | 316 | | Kegg ID | C00128 | | ChemSpider ID | 395082 | | Wikipedia ID | Not Available | | BioCyc ID | Not Available | | Ligand Expo | NCC |
|
|---|