| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-08-03 14:57:55 -0600 |
|---|
| Update Date | 2015-09-13 12:56:16 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | 2-Aminoisobutyric acid |
|---|
| Description | 2-Aminoisobutyric acid (or AIB) is a nonprotein amino acid that is found in certain antibiotics. It can be used as a nitrogen source by certain soil bacteria. AIB may also be produced as an end-product of pyrimidine metabolism. This compound has been identified in metabolomic experiments on E. coli. |
|---|
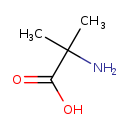
| Structure | |
|---|
| Synonyms: | - 2-Amino-2-methylpropanoate
- 2-Amino-2-methylpropanoic acid
- 2-Aminoisobutyrate
- 2-Methylalanine
- A-Aminoisobutanoate
- A-Aminoisobutanoic acid
- A-Aminoisobutyrate
- A-Aminoisobutyric acid
- A-Methylalanine
- AIB
- Alpha-Aminoisobutanoate
- Alpha-Aminoisobutanoic acid
- Alpha-Aminoisobutyrate
- Alpha-Aminoisobutyric acid
- Alpha-Methylalanine
- α-Aminoisobutanoate
- α-Aminoisobutanoic acid
- α-Aminoisobutyrate
- α-Aminoisobutyric acid
- α-Methylalanine
|
|---|
| Chemical Formula: | C4H9NO2 |
|---|
| Weight: | Average: 103.1198
Monoisotopic: 103.063328537 |
|---|
| InChI Key: | FUOOLUPWFVMBKG-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C4H9NO2/c1-4(2,5)3(6)7/h5H2,1-2H3,(H,6,7) |
|---|
| CAS number: | 62-57-7 |
|---|
| IUPAC Name: | 2-amino-2-methylpropanoic acid |
|---|
| Traditional IUPAC Name: | α-aminoisobutyric acid |
|---|
| SMILES: | CC(C)(N)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alpha amino acids. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Alpha amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-amino acid
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Organic oxide
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Primary aliphatic amine
- Organic oxygen compound
- Carbonyl group
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | 0 |
|---|
| Melting point: | 335 °C |
|---|
| Experimental Properties: | | Property | Value | Source |
|---|
| Water Solubility: | 181 mg/mL at 25 oC [YALKOWSKY,SH & DANNENFELSER,RM (1992)] | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| | Concentration | Strain | Media | Growth Status | Growth System | Temperature | Details |
|---|
| 11± 0 uM | BW25113 | 48 mM Na2HPO4, 22 mM KH2PO4, 10 mM NaCl, 45 mM (NH4)2SO4, supplemented with 1 mM MgSO4, 1 mg/l thiamine·HCl, 5.6 mg/l CaCl2, 8 mg/l FeCl3, 1 mg/l MnCl2·4H2O, 1.7 mg/l ZnCl2, 0.43 mg/l CuCl2·2H2O, 0.6 mg/l CoCl2·2H2O and 0.6 mg/l Na2MoO4·2H2O. 4 g/L Gluco | Stationary Phase, glucose limited | Bioreactor, pH controlled, O2 and CO2 controlled, dilution rate: 0.2/h | 37 oC | PMID: 17379776 |
|
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-001i-0900000000-e469c31fdb361c68d7c1 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (2 TMS) | splash10-001i-0900000000-a36ad39a36ae444e46ae | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Non-derivatized) | splash10-001i-0900000000-e469c31fdb361c68d7c1 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (Non-derivatized) | splash10-001i-0900000000-a36ad39a36ae444e46ae | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-9000000000-6b7bf93c06f2aeefce39 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-9600000000-a300b153080dc92a72f6 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-000i-9200000000-da2827369e92adba7cd8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0udi-7900000000-6079ac9d44cf4a311152 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0udi-4900000000-cf4a983a808aa8a18729 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0udi-0900000000-b09f9465bfd507f59a9d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0udi-0900000000-4eeda03ca057f971ad21 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0udi-4900000000-8b7badd6f8457356fe62 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-0a4i-9000000000-7e17058c19f5affb357a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-0a4i-9000000000-b986c9a42c2c739c6fd0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-0a4i-9000000000-bf285168a5a5d41ab049 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-0a4l-9000000000-b7fd609f2bf6e6145d54 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0pb9-9500000000-2f9ed721c06843a9df2b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0udi-0900000000-02624b93137883b214ad | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0udi-0900000000-b09f9465bfd507f59a9d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0udi-0900000000-4eeda03ca057f971ad21 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0udi-0900000000-02624b93137883b214ad | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , negative | splash10-0udi-0900000000-aac342be2df7745c885d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0udi-4900000000-8b7badd6f8457356fe62 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0a4i-9000000000-23a671f383f99bc465c8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0a4i-9000000000-b986c9a42c2c739c6fd0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-9200000000-4849dde814173ad8e73f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9000000000-5ae41491e22202d700f3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9000000000-6dff5b04f9cf69c4d686 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-278a45a568ba95d65cab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-1900000000-82d99c3456fa9229a224 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0k9i-9200000000-a1add3dee96321589e43 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - CRUMPLER HR, DENT CE, HARRIS H, WESTALL RG: beta-Aminoisobutyric acid (alpha-methyl-beta-alanine); a new amino-acid obtained from human urine. Nature. 1951 Feb 24;167(4243):307-8. Pubmed: 14806475
- HARRIS H: Family studies on the urinary excretion of beta-aminoisobutyric acid. Ann Eugen. 1953 Jun;18(1):43-9. Pubmed: 13058271
- Ishii, N., Nakahigashi, K., Baba, T., Robert, M., Soga, T., Kanai, A., Hirasawa, T., Naba, M., Hirai, K., Hoque, A., Ho, P. Y., Kakazu, Y., Sugawara, K., Igarashi, S., Harada, S., Masuda, T., Sugiyama, N., Togashi, T., Hasegawa, M., Takai, Y., Yugi, K., Arakawa, K., Iwata, N., Toya, Y., Nakayama, Y., Nishioka, T., Shimizu, K., Mori, H., Tomita, M. (2007). "Multiple high-throughput analyses monitor the response of E. coli to perturbations." Science 316:593-597. Pubmed: 17379776
- Lucarelli C, Betto P, Ricciarello G, Grossi G: High-performance liquid chromatographic determination of L-3-(3,4-dihydroxyphenyl)-2-methylalanine (alpha-methyldopa) in human urine and plasma. J Chromatogr. 1991 Mar 22;541(1-2):285-96. Pubmed: 2037651
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. Pubmed: 19212411
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| Links |
|---|
| External Links: | |
|---|