| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-07-30 14:55:41 -0600 |
|---|
| Update Date | 2015-09-13 12:56:15 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
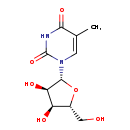
| Name: | Ribothymidine |
|---|
| Description | Ribothymidine is a pyrimidine nucleoside that is also known as 5-methyluridine. (Wikipedia) It is a methylated version of uridine, one of the building blocks of RNA. It also differs from thymidine, one of the building blocks of DNA, by having an extra hydroxyl (OH group) on its ribose ring. (inferred from compound structure) |
|---|
| Structure | |
|---|
| Synonyms: | - 1-(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl-5-methylpyrimidine-2,4-dione
- 1-(b-D-Ribofuranosyl)thymine
- 1-(beta-D-ribofuranosyl)thymine
- 1-(β-D-Ribofuranosyl)thymine
- 1-b-D-Ribofuranosylthymine
- 1-b-delta-Ribofuranosylthymine
- 1-b-δ-Ribofuranosylthymine
- 1-beta-delta-Ribofuranosylthymine
- 1-β-δ-Ribofuranosylthymine
- 5-Methyl-1-b-D-ribofuranosyl-2,4(1H,3H)-pyrimidinedione
- 5-Methyl-1-b-delta-ribofuranosyl-2,4(1H,3H)-pyrimidinedione
- 5-Methyl-1-b-δ-ribofuranosyl-2,4(1H,3H)-pyrimidinedione
- 5-methyl-1-beta-D-ribofuranosyl-2,4(1H,3H)-Pyrimidinedione
- 5-methyl-1-beta-delta-ribofuranosyl-2,4(1H,3H)-Pyrimidinedione
- 5-Methyl-1-β-D-ribofuranosyl-2,4(1H,3H)-pyrimidinedione
- 5-Methyl-1-β-δ-ribofuranosyl-2,4(1H,3H)-pyrimidinedione
- 5-methyl-Uridine
- 5-Methyluridine
- b-D-Ribofuranoside
- B-D-Ribofuranoside thymine-1
- b-delta-Ribofuranoside
- b-delta-Ribofuranoside thymine-1
- b-δ-Ribofuranoside
- b-δ-Ribofuranoside thymine-1
- Beta-D-Ribofuranoside
- Beta-delta-Ribofuranoside
- Beta-delta-Ribofuranoside thymine-1
- Ribosylthymidine
- Ribosylthymine
- T
- Thymine ribofuranoside
- Thymine ribonucleoside
- Thymine riboside
- Thymine-1 b-D-ribofuranosylthymine
- Thymine-1 b-delta-ribofuranosylthymine
- Thymine-1 b-δ-ribofuranosylthymine
- Thymine-1 beta-D-Ribofuranosylthymine
- Thymine-1 beta-delta-Ribofuranosylthymine
- Thymine-1 β-D-ribofuranosylthymine
- Thymine-1 β-δ-ribofuranosylthymine
- β-D-Ribofuranoside
- β-δ-Ribofuranoside
- β-δ-Ribofuranoside thymine-1
|
|---|
| Chemical Formula: | C10H14N2O6 |
|---|
| Weight: | Average: 258.228
Monoisotopic: 258.08518619 |
|---|
| InChI Key: | DWRXFEITVBNRMK-JXOAFFINSA-N |
|---|
| InChI: | InChI=1S/C10H14N2O6/c1-4-2-12(10(17)11-8(4)16)9-7(15)6(14)5(3-13)18-9/h2,5-7,9,13-15H,3H2,1H3,(H,11,16,17)/t5-,6-,7-,9-/m1/s1 |
|---|
| CAS number: | 1463-10-1 |
|---|
| IUPAC Name: | 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-5-methyl-1,2,3,4-tetrahydropyrimidine-2,4-dione |
|---|
| Traditional IUPAC Name: | thymidin |
|---|
| SMILES: | CC1=CN([C@@H]2O[C@H](CO)[C@@H](O)[C@H]2O)C(=O)NC1=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine nucleosides. Pyrimidine nucleosides are compounds comprising a pyrimidine base attached to a ribosyl or deoxyribosyl moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleosides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Pyrimidine nucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine nucleoside
- Glycosyl compound
- N-glycosyl compound
- Pentose monosaccharide
- Pyrimidone
- Hydropyrimidine
- Monosaccharide
- Pyrimidine
- Vinylogous amide
- Tetrahydrofuran
- Heteroaromatic compound
- Urea
- Secondary alcohol
- Lactam
- Organoheterocyclic compound
- Oxacycle
- Azacycle
- Hydrocarbon derivative
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Primary alcohol
- Alcohol
- Organic oxygen compound
- Organic oxide
- Organic nitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | 0 |
|---|
| Melting point: | 183-187 °C |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05bf-9420000000-897dab8f4bbf7b8e664c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0kp0-6879400000-656a0a43be4a2cd044e7 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-004i-0900000000-1653ad80c56ae3727f92 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-004i-2900000000-826774188d3e78ca38f2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a6r-9800000000-f5daf31a98b364b70f8c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0090000000-f186cfbb52edc03ff4de | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-004i-0900000000-9f7398feab5dd8950264 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-893019db60499c0a9602 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-004i-2900000000-461a9c94bf89e5b47970 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0900000000-1db6a34eff49b044d944 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-004i-1900000000-1c2ee21cd9d3515f7124 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-056r-1970000000-ba6f9aa1164c365abfa2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-01u0-8900000000-a63ffd4c990b9c9a543a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-004i-4900000000-71ebad0df798bf2ac282 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-06w9-8900000000-3c0ceb68319de86fc30f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-004i-1900000000-45b9a7b5e831351a1fe0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0900000000-e1eddf8b44309a38eebb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0910000000-90f6da130f49fa9df29d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-5900000000-203a4367756530615ffd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004j-9800000000-13e51659da01285bdc7f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a6r-1490000000-4ba707a2228996f9f9f7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-6920000000-a25c60ade61d6d32f0f1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001m-9100000000-37e601ae2d82d72111e7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0900000000-f08754aed5978a645b7c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-3910000000-01ae005a14dd4c188d46 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-9700000000-513e87b6fba58d79ef98 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-056r-0950000000-b4a8bf9fcddaedc20b74 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Avraham Y, Grossowicz N, Yashphe J: Purification and characterization of uridine and thymidine phosphorylase from Lactobacillus casei. Biochim Biophys Acta. 1990 Sep 3;1040(2):287-93. Pubmed: 2119230
- Cho SH, Jung BH, Lee SH, Lee WY, Kong G, Chung BC: Direct determination of nucleosides in the urine of patients with breast cancer using column-switching liquid chromatography-tandem mass spectrometry. Biomed Chromatogr. 2006 Nov;20(11):1229-36. Pubmed: 16799933
- Hacia JG, Woski SA, Fidanza J, Edgemon K, Hunt N, McGall G, Fodor SP, Collins FS: Enhanced high density oligonucleotide array-based sequence analysis using modified nucleoside triphosphates. Nucleic Acids Res. 1998 Nov 1;26(21):4975-82. Pubmed: 9776762
- Lang TT, Selner M, Young JD, Cass CE: Acquisition of human concentrative nucleoside transporter 2 (hcnt2) activity by gene transfer confers sensitivity to fluoropyrimidine nucleosides in drug-resistant leukemia cells. Mol Pharmacol. 2001 Nov;60(5):1143-52. Pubmed: 11641443
- Liu Z, Liu S, Xie Z, Blum W, Perrotti D, Paschka P, Klisovic R, Byrd J, Chan KK, Marcucci G: Characterization of in vitro and in vivo hypomethylating effects of decitabine in acute myeloid leukemia by a rapid, specific and sensitive LC-MS/MS method. Nucleic Acids Res. 2007;35(5):e31. Epub 2007 Jan 30. Pubmed: 17264127
- Mori T, Guo MW, Li X, Xu JP, Mori E: Isolation and identification of apoptosis inducing nucleosides from CD57(+)HLA-DRbright natural suppressor cell line. Biochem Biophys Res Commun. 1998 Oct 20;251(2):416-22. Pubmed: 9792789
- Morris GS, Simmonds HA, Davies PM: Use of biological fluids for the rapid diagnosis of potentially lethal inherited disorders of human purine and pyrimidine metabolism. Biomed Chromatogr. 1986 Jun;1(3):109-18. Pubmed: 3506820
- Urbonavicius J, Durand JM, Bjork GR: Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri. J Bacteriol. 2002 Oct;184(19):5348-57. Pubmed: 12218021
- Weisbart RH, Garrett RA, Liebling MR, Barnett EV, Paulus HE, Katz DH: Specificity of anti-nucleoside antibodies in systemic lupus erythematosus. Clin Immunol Immunopathol. 1983 Jun;27(3):403-11. Pubmed: 6603316
- Yang TH, Hu ML: Intracellular levels of S-adenosylhomocysteine but not homocysteine are highly correlated to the expression of nm23-H1 and the level of 5-methyldeoxycytidine in human hepatoma cells with different invasion activities. Nutr Cancer. 2006;55(2):224-31. Pubmed: 17044778
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| Links |
|---|
| External Links: | |
|---|