| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-07-30 14:55:41 -0600 |
|---|
| Update Date | 2015-09-13 15:15:32 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
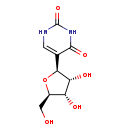
| Name: | Pseudouridine |
|---|
| Description | Pseudouridine is the C-glycoside isomer of the nucleoside uridine, and it is the most prevalent of the over one hundred different modified nucleosides found in RNA. Pseudouridine is found in all species and in all classes of RNA except mRNA. It is formed by enzymes called pseudouridine synthases, which post-transcriptionally isomerize specific uridine residues in RNA (Wikipedia). |
|---|
| Structure | |
|---|
| Synonyms: | - (1S)-1,4-anhydro-1-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-D-ribitol
- 5-(2S,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl-1H-pyrimidine-2,4-dione
- 5-(b-D-Ribofuranosyl)uracil
- 5-(b-delta-Ribofuranosyl)uracil
- 5-(b-δ-Ribofuranosyl)uracil
- 5-(beta-D-ribofuranosyl)uracil
- 5-(β-D-Ribofuranosyl)uracil
- 5-b-D-ribofuranosyl-Uracil
- 5-b-delta-Ribofuranosyl-uracil
- 5-b-δ-Ribofuranosyl-uracil
- 5-beta-delta-ribofuranosyl-Uracil
- 5-Ribosyluracil
- 5-β-δ-Ribofuranosyl-uracil
- B-D-Pseudouridine
- b-delta-Pseudouridine
- B-Pseudouridine
- b-δ-Pseudouridine
- Beta-delta-Pseudouridine
- Beta-Pseudouridine
- P
- Pseudouridine C
- Psi-uridine
- Y-Uridine
- β-Pseudouridine
- β-δ-Pseudouridine
|
|---|
| Chemical Formula: | C9H12N2O6 |
|---|
| Weight: | Average: 244.2014
Monoisotopic: 244.069536126 |
|---|
| InChI Key: | PTJWIQPHWPFNBW-GBNDHIKLSA-N |
|---|
| InChI: | InChI=1S/C9H12N2O6/c12-2-4-5(13)6(14)7(17-4)3-1-10-9(16)11-8(3)15/h1,4-7,12-14H,2H2,(H2,10,11,15,16)/t4-,5-,6-,7+/m1/s1 |
|---|
| CAS number: | 1445-07-4 |
|---|
| IUPAC Name: | 5-[(2S,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2,3,4-tetrahydropyrimidine-2,4-dione |
|---|
| Traditional IUPAC Name: | β-pseudouridine |
|---|
| SMILES: | OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)C1=CNC(=O)NC1=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as nucleoside and nucleotide analogues. These are analogues of nucleosides and nucleotides. These include phosphonated nucleosides, C-glycosylated nucleoside bases, analogues where the sugar unit is a pyranose, and carbocyclic nucleosides, among others. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Nucleoside and nucleotide analogues |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Nucleoside and nucleotide analogues |
|---|
| Alternative Parents | |
|---|
| Substituents | - C-glycosyl compound
- Glycosyl compound
- Pentose monosaccharide
- Pyrimidone
- Hydropyrimidine
- Monosaccharide
- Pyrimidine
- Heteroaromatic compound
- Tetrahydrofuran
- Vinylogous amide
- Lactam
- Secondary alcohol
- Urea
- Azacycle
- Oxacycle
- Ether
- Dialkyl ether
- Organoheterocyclic compound
- Primary alcohol
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Alcohol
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | 0 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0unc-7930000000-a1e7f343c88a63ac1976 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0fi0-3494100000-8d726ed412554c1f92f0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-0udi-0900000000-6ade42aaa885371e176c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 10V, positive | splash10-0a4i-0190000000-129f123affa0ecbf7a59 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 20V, positive | splash10-052f-0910000000-302fbdef606070bd5a67 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 40V, positive | splash10-001i-9300000000-11f5761be16d1b061df5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-0006-9600000000-7585746143c62ab3f832 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-010da2c57a936868eecc | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0kai-9200000000-12f986238bdf05dc39b9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-001i-9400000000-b6dbdfa2d4643fc13e70 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0190000000-129f123affa0ecbf7a59 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-052f-0910000000-302fbdef606070bd5a67 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 0V, Positive | splash10-0bta-0890000000-2b3810ae2bbe68268b17 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-001i-9300000000-7e64dc8404ac65411358 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 0V, Positive | splash10-052b-0290000000-7cf7d5be6f7a84299cab | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0950000000-de2a45f204636756b9c4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-00l6-9000000000-b4060d47530bdf70d6cc | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-3920000000-15bb158a77906d0f87b5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-2900000000-600643d24abd3ba9b5f1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0920000000-a73049fd7001db7f72d5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0900000000-46e1b064445ad43a60fd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0490000000-226d044c24dd80a7c2ca | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0h0s-2980000000-add34ee37b95c102114d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-06rm-5900000000-8d66d80928e84e9c93fd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9360000000-3bd8fcdd66c5d245778e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9210000000-f551740a13c90ff1d324 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-bf4e4234a47d8aa03828 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Bernert JT Jr, Bell CJ, Guntupalli J, Hannon WH: Pseudouridine is unsuitable as an endogenous renal clearance marker. Clin Chem. 1988 Jun;34(6):1011-7. Pubmed: 3378317
- Colonna A, Russo T, Esposito F, Salvatore F, Cimino F: Determination of pseudouridine and other nucleosides in human blood serum by high-performance liquid chromatography. Anal Biochem. 1983 Apr 1;130(1):19-26. Pubmed: 6869800
- Lee SH, Jung BH, Kim SY, Chung BC: A rapid and sensitive method for quantitation of nucleosides in human urine using liquid chromatography/mass spectrometry with direct urine injection. Rapid Commun Mass Spectrom. 2004;18(9):973-7. Pubmed: 15116424
- Mak TW, Ho SS, Ho CS, Jones MG, Lai CK, Lam CW: Pleural fluid pseudouridine in malignant and benign pleural effusions. Ann Clin Biochem. 1998 Jan;35 ( Pt 1):94-8. Pubmed: 9463745
- Shoemaker JD, Elliott WH: Automated screening of urine samples for carbohydrates, organic and amino acids after treatment with urease. J Chromatogr. 1991 Jan 2;562(1-2):125-38. Pubmed: 2026685
- Uziel M, Smith LH, Taylor SA: Modified nucleosides in urine: selective removal and analysis. Clin Chem. 1976 Sep;22(9):1451-5. Pubmed: 954194
- Woodcock TM, Chou TC, Tan CT, Sternberg SS, Philips FS, Young CW, Burchenal JH: Biochemical, pharmacological, and phase I clinical evaluation of pseudoisocytidine. Cancer Res. 1980 Nov;40(11):4243-9. Pubmed: 7471064
|
|---|
| Synthesis Reference: | anessian, Stephen; Machaalani, Roger. A highly stereo-controlled and efficient synthesis of a- and b-pseudouridines. Tetrahedron Letters (2003), 44(45), 8321-8323. |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|