Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose (M2MDB001762)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-07-30 14:55:30 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:21:14 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose is a member of the chemical class known as Pyrimidine Nucleotide Sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
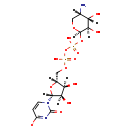
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C14H22N3O15P2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 534.2831 Monoisotopic: 534.052615073 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | GWBAKYBSWHQNMQ-IAZOVDBXSA-M | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C14H23N3O15P2/c15-5-3-28-13(11(22)8(5)19)31-34(26,27)32-33(24,25)29-4-6-9(20)10(21)12(30-6)17-2-1-7(18)16-14(17)23/h1-2,5-6,8-13,19-22H,3-4,15H2,(H,24,25)(H,26,27)(H,16,18,23)/p-1/t5-,6+,8-,9+,10+,11+,12+,13+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 1-[(2R,3R,4S,5R)-5-[({[({[(2R,3R,4S,5S)-5-amino-3,4-dihydroxyoxan-2-yl]oxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-3,4-dihydroxyoxolan-2-yl]-2-oxo-1,2-dihydropyrimidin-4-olate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 1-[(2R,3R,4S,5R)-5-{[({[(2R,3R,4S,5S)-5-amino-3,4-dihydroxyoxan-2-yl]oxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxy]methyl}-3,4-dihydroxyoxolan-2-yl]-2-oxopyrimidin-4-olate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@]1(COP(O)(=O)OP(O)(=O)O[C@@]2([H])OC[C@]([H])(N)[C@]([H])(O)[C@@]2([H])O)O[C@@]([H])(N2C=CC([O-])=NC2=O)[C@]([H])(O)[C@]1([H])O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as pyrimidine ribonucleoside diphosphates. These are pyrimidine ribonucleotides with diphosphate group linked to the ribose moiety. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Pyrimidine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Pyrimidine ribonucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pyrimidine ribonucleoside diphosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | L-Glutamate + UDP-4-Keto-pyranose <> alpha-Ketoglutarate + Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose N10-Formyl-THF + Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose <> Hydrogen ion + Tetrahydrofolic acid + Uridine 5''-diphospho-{beta}-4-deoxy-4-formamido-L-arabinose N10-Formyl-THF + Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose <> Tetrahydrofolic acid + Uridine 5''-diphospho-{beta}-4-deoxy-4-formamido-L-arabinose Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose + N10-Formyl-THF > Hydrogen ion + Uridine 5''-diphospho-{beta}-4-deoxy-4-formamido-L-arabinose + Tetrahydrofolic acid Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose + Oxoglutaric acid <> UDP-4-Keto-pyranose + L-Glutamate Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose + an N10-formyl-tetrahydrofolate + N10-Formyl-THF > UDP-4-Deoxy-4-formamido-beta-L-arabinose + Hydrogen ion + a tetrahydrofolate + Tetrahydrofolic acid UDP-β-L-threo-pentapyranos-4-ulose + L-Glutamic acid + L-Glutamate > Oxoglutaric acid + Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose L-Glutamate + UDP-4-Keto-pyranose <> alpha-Ketoglutarate + Uridine 5''-diphospho-{beta}-4-deoxy-4-amino-L-arabinose | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in hydroxymethyl-, formyl- and related transferase activity
- Specific function:
- Bifunctional enzyme that catalyzes the oxidative decarboxylation of UDP-glucuronic acid (UDP-GlcUA) to UDP-4-keto- arabinose (UDP-Ara4O) and the addition of a formyl group to UDP-4- amino-4-deoxy-L-arabinose (UDP-L-Ara4N) to form UDP-L-4-formamido- arabinose (UDP-L-Ara4FN). The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides
- Gene Name:
- arnA
- Uniprot ID:
- P77398
- Molecular weight:
- 74288
Reactions
| UDP-alpha-D-glucuronate + NAD(+) = UDP-beta-L-threo-pentapyranos-4-ulose + CO(2) + NADH. |
| 10-formyltetrahydrofolate + UDP-4-amino-4-deoxy-beta-L-arabinose = 5,6,7,8-tetrahydrofolate + UDP-4-deoxy-4-formamido-beta-L-arabinose. |
- General function:
- Involved in transaminase activity
- Specific function:
- Catalyzes the conversion of UDP-4-keto-arabinose (UDP- Ara4O) to UDP-4-amino-4-deoxy-L-arabinose (UDP-L-Ara4N). The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides
- Gene Name:
- arnB
- Uniprot ID:
- P77690
- Molecular weight:
- 41649
Reactions
| UDP-4-amino-4-deoxy-beta-L-arabinose + 2-oxoglutarate = UDP-beta-L-threo-pentapyranos-4-ulose + glutamate. |