Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Butanesulfonate (M2MDB001715)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-07-30 14:55:17 -0600 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:21:08 -0600 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Butanesulfonate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Butanesulfonate is a member of the chemical class known as Sulfonic Acids. These are compounds containing the sulfonic acid group, which has the general structure RS(=O)2OH (R = H). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
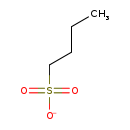
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C4H9O3S | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 137.177 Monoisotopic: 137.027239844 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | QDHFHIQKOVNCNC-UHFFFAOYSA-M | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C4H10O3S/c1-2-3-4-8(5,6)7/h2-4H2,1H3,(H,5,6,7)/p-1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | butane-1-sulfonate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 1-butanesulfonate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CCCCS([O-])(=O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as organosulfonic acids. Organosulfonic acids are compounds containing the sulfonic acid group, which has the general structure RS(=O)2OH (R is not a hydrogen atom). | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organic sulfonic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Organosulfonic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Organosulfonic acids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + Butanesulfonate > ADP + Butanesulfonate + Hydrogen ion + Phosphate Adenosine triphosphate + Water + Butanesulfonate > ADP + Butanesulfonate + Hydrogen ion + Phosphate Butanesulfonate + FMNH + Oxygen > Butanal + Flavin Mononucleotide + Hydrogen ion + Water + Sulfite Butanesulfonate + Oxygen + FMNH > Butanal + Sulfite + Water + Flavin Mononucleotide + Hydrogen ion Butanesulfonate + Oxygen + FMNH2 > Hydrogen ion + Water + Sulfite + Flavin Mononucleotide + Betaine aldehyde + Sulfite Adenosine triphosphate + Water + Butanesulfonate > Adenosine diphosphate + Phosphate + Hydrogen ion + Butanesulfonate + ADP Adenosine triphosphate + Water + Butanesulfonate > Adenosine diphosphate + Phosphate + Hydrogen ion + Butanesulfonate + ADP | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in alkanesulfonate monooxygenase activity
- Specific function:
- Involved in desulfonation of aliphatic sulfonates. Catalyzes the conversion of pentanesulfonic acid to sulfite and pentaldehyde and is able to desulfonate a wide range of sulfonated substrates including C-2 to C-10 unsubstituted linear alkanesulfonates, substituted ethanesulfonic acids and sulfonated buffers
- Gene Name:
- ssuD
- Uniprot ID:
- P80645
- Molecular weight:
- 41736
Reactions
| An alkanesufonate (R-CH(2)-SO(3)H) + FMNH(2) + O(2) = an aldehyde (R-CHO) + FMN + sulfite + H(2)O. |
- General function:
- Involved in transporter activity
- Specific function:
- Part of a binding-protein-dependent transport system for aliphatic sulfonates. Probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- ssuC
- Uniprot ID:
- P75851
- Molecular weight:
- 28925
- General function:
- Involved in transporter activity
- Specific function:
- Part of a binding-protein-dependent transport system for taurine. Probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- tauC
- Uniprot ID:
- Q47539
- Molecular weight:
- 29812
- General function:
- Involved in sulfur compound metabolic process
- Specific function:
- Part of a binding-protein-dependent transport system for aliphatic sulfonates. Putative binding protein
- Gene Name:
- ssuA
- Uniprot ID:
- P75853
- Molecular weight:
- 34557
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex TauABC involved in taurine import. Responsible for energy coupling to the transport system
- Gene Name:
- tauB
- Uniprot ID:
- Q47538
- Molecular weight:
- 28297
Reactions
| ATP + H(2)O + taurine(Out) = ADP + phosphate + taurine(In). |
- General function:
- Involved in transporter activity
- Specific function:
- Part of a binding-protein-dependent transport system for taurine
- Gene Name:
- tauA
- Uniprot ID:
- Q47537
- Molecular weight:
- 34266
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex SsuABC involved in aliphatic sulfonates import. Responsible for energy coupling to the transport system (Probable)
- Gene Name:
- ssuB
- Uniprot ID:
- P0AAI1
- Molecular weight:
- 27738
Transporters
- General function:
- Involved in transporter activity
- Specific function:
- Part of a binding-protein-dependent transport system for aliphatic sulfonates. Probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- ssuC
- Uniprot ID:
- P75851
- Molecular weight:
- 28925
- General function:
- Involved in transporter activity
- Specific function:
- Part of a binding-protein-dependent transport system for taurine. Probably responsible for the translocation of the substrate across the membrane
- Gene Name:
- tauC
- Uniprot ID:
- Q47539
- Molecular weight:
- 29812
- General function:
- Involved in sulfur compound metabolic process
- Specific function:
- Part of a binding-protein-dependent transport system for aliphatic sulfonates. Putative binding protein
- Gene Name:
- ssuA
- Uniprot ID:
- P75853
- Molecular weight:
- 34557
- General function:
- Involved in transporter activity
- Specific function:
- Non-specific porin
- Gene Name:
- ompN
- Uniprot ID:
- P77747
- Molecular weight:
- 41220
- General function:
- Involved in transporter activity
- Specific function:
- Uptake of inorganic phosphate, phosphorylated compounds, and some other negatively charged solutes
- Gene Name:
- phoE
- Uniprot ID:
- P02932
- Molecular weight:
- 38922
- General function:
- Involved in transporter activity
- Specific function:
- OmpF is a porin that forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane. It is also a receptor for the bacteriophage T2
- Gene Name:
- ompF
- Uniprot ID:
- P02931
- Molecular weight:
- 39333
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex TauABC involved in taurine import. Responsible for energy coupling to the transport system
- Gene Name:
- tauB
- Uniprot ID:
- Q47538
- Molecular weight:
- 28297
Reactions
| ATP + H(2)O + taurine(Out) = ADP + phosphate + taurine(In). |
- General function:
- Involved in transporter activity
- Specific function:
- Part of a binding-protein-dependent transport system for taurine
- Gene Name:
- tauA
- Uniprot ID:
- Q47537
- Molecular weight:
- 34266
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex SsuABC involved in aliphatic sulfonates import. Responsible for energy coupling to the transport system (Probable)
- Gene Name:
- ssuB
- Uniprot ID:
- P0AAI1
- Molecular weight:
- 27738
- General function:
- Involved in transporter activity
- Specific function:
- Forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane
- Gene Name:
- ompC
- Uniprot ID:
- P06996
- Molecular weight:
- 40368