Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
UDP-N-Acetyl-D-mannosaminouronate (M2MDB001698)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-07-30 14:55:15 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:21:06 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | UDP-N-Acetyl-D-mannosaminouronate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | UDP-n-acetyl-D-mannosaminouronate is a member of the chemical class known as Pyrimidine Nucleotide Sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
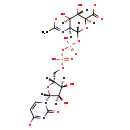
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C17H22N3O18P2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 618.3134 Monoisotopic: 618.037358939 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | DZOGQXKQLXAPND-XHUKORKBSA-K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C17H25N3O18P2/c1-5(21)18-8-10(24)11(25)13(15(27)28)36-16(8)37-40(32,33)38-39(30,31)34-4-6-9(23)12(26)14(35-6)20-3-2-7(22)19-17(20)29/h2-3,6,8-14,16,23-26H,4H2,1H3,(H,18,21)(H,27,28)(H,30,31)(H,32,33)(H,19,22,29)/p-3/t6-,8+,9-,10-,11+,12-,13+,14-,16-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S,3S,4R,5S,6R)-6-({[({[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-oxido-2-oxo-1,2-dihydropyrimidin-1-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-3,4-dihydroxy-5-[(1-oxidoethylidene)amino]oxane-2-carboxylate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (2S,3S,4R,5S,6R)-6-[({[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-oxido-2-oxopyrimidin-1-yl)oxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxy]-3,4-dihydroxy-5-[(1-oxidoethylidene)amino]oxane-2-carboxylate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@]1(COP(O)(=O)OP(O)(=O)O[C@@]2([H])O[C@]([H])(C([O-])=O)[C@@]([H])(O)[C@]([H])(O)[C@]2([H])N=C(C)[O-])O[C@@]([H])(N2C=CC([O-])=NC2=O)[C@]([H])(O)[C@]1([H])O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as pyrimidine nucleotide sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Pyrimidine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Pyrimidine nucleotide sugars | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pyrimidine nucleotide sugars | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Water + 2 NAD + UDP-N-Acetyl-D-mannosamine <>3 Hydrogen ion +2 NADH + UDP-N-Acetyl-D-mannosaminouronate UDP-N-Acetyl-D-mannosaminouronate + Undecaprenyl-N-acetyl-alpha-D-glucosaminyl-pyrophosphate > Hydrogen ion + Uridine 5'-diphosphate + Undecaprenyl-diphospho-N-acetylglucosamine-N-acetylmannosaminuronate UDP-N-Acetyl-D-mannosamine + 2 NAD + Water <> UDP-N-Acetyl-D-mannosaminouronate +2 NADH +2 Hydrogen ion Undecaprenyl-N-acetyl-alpha-D-glucosaminyl-pyrophosphate + UDP-N-Acetyl-D-mannosaminouronate <> Hydrogen ion + Undecaprenyl phosphate + Uridine 5'-diphosphate UDP-N-Acetyl-D-mannosamine + NAD + Water <> UDP-N-Acetyl-D-mannosaminouronate + NADH + Hydrogen ion UDP-N-Acetyl-D-mannosamine + 2 NAD + Water > UDP-N-Acetyl-D-mannosaminouronate +2 NADH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in oxidation-reduction process
- Specific function:
- Catalyzes the four-electron oxidation of UDP-N-acetyl-D- mannosamine (UDP-ManNAc), reducing NAD(+) and releasing UDP-N- acetylmannosaminuronic acid (UDP-ManNAcA)
- Gene Name:
- wecC
- Uniprot ID:
- P27829
- Molecular weight:
- 45838
Reactions
| UDP-N-acetyl-D-mannosamine + 2 NAD(+) + H(2)O = UDP-N-acetyl-D-mannosaminuronate + 2 NADH. |
- General function:
- Involved in transferase activity, transferring hexosyl groups
- Specific function:
- Catalyzes the synthesis of Und-PP-GlcNAc-ManNAcA (Lipid II), the second lipid-linked intermediate involved in ECA synthesis
- Gene Name:
- wecG
- Uniprot ID:
- P27836
- Molecular weight:
- 27928
Reactions
| UDP-ManNAcA + Und-PP-GlcNAc = UDP + Und-PP-GlcNAc-ManNAcA. |