| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-07-30 14:55:11 -0600 |
|---|
| Update Date | 2015-06-03 17:21:04 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Phosphotyrosine |
|---|
| Description | Phosphotyrosine is a tyrosine (an amino acid) with a phosphate attached to its aromatic ring. Tyrosine (abbreviated as Tyr or Y) is one of the 22 amino acids that are used by cells to synthesize proteins. It is a non-essential amino acid with a polar side group. (Wikipedia) Tyrosine phosphorylation and dephosphorylation plays a role in cellular signal transduction and possibly in cell growth control. (PubChem) In E. coli, phosphotyrosine can act as a noncompetitive inhibitor of the enzyme biosynthetic ornithine decarboxylase. (EcoCyc) |
|---|
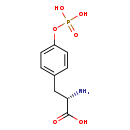
| Structure | |
|---|
| Synonyms: | - (S)-2-amino-3-(4-hydroxyphenyl)propanoate 4'-phosphate
- (S)-2-amino-3-(4-hydroxyphenyl)propanoic acid 4'-phosphate
- (S)-2-amino-3-(4-Hydroxyphenyl)propanoic acid 4'-phosphoric acid
- L-3-(4-Hydroxyphenyl)alanine 4'-phosphate
- L-3-(4-Hydroxyphenyl)alanine 4'-phosphoric acid
- L-Phosphotyrosine
- L-Tyrosine, dihydrogen phosphate (ester)
- L-Tyrosine, dihydrogen phosphoric acid (ester)
- L-Tyrosine-O-phosphate
- L-Tyrosine-O-phosphoric acid
- O-Phospho-L-tyrosine
- O-Phosphono-L-tyrosine
- O-Phosphotyrosine
- O4'-phospho-L-tyrosine
- O4-phosphono-L-tyrosine
- O4-phosphotyrosine
- Phospho-L-tyrosine
- Phosphonotyrosine
- Phosphotyrosine
- Phosphotyrosine (pY)
- Tyrosine O-phosphate
- Tyrosine O-phosphoric acid
- Tyrosine phosphate
- Tyrosine phosphoric acid
|
|---|
| Chemical Formula: | C9H12NO6P |
|---|
| Weight: | Average: 261.1684
Monoisotopic: 261.040223633 |
|---|
| InChI Key: | DCWXELXMIBXGTH-QMMMGPOBSA-N |
|---|
| InChI: | InChI=1S/C9H12NO6P/c10-8(9(11)12)5-6-1-3-7(4-2-6)16-17(13,14)15/h1-4,8H,5,10H2,(H,11,12)(H2,13,14,15)/t8-/m0/s1 |
|---|
| CAS number: | 21820-51-9 |
|---|
| IUPAC Name: | (2S)-2-amino-3-[4-(phosphonooxy)phenyl]propanoic acid |
|---|
| Traditional IUPAC Name: | phosphonotyrosine |
|---|
| SMILES: | N[C@@H](CC1=CC=C(OP(O)(O)=O)C=C1)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phenylalanine and derivatives. Phenylalanine and derivatives are compounds containing phenylalanine or a derivative thereof resulting from reaction of phenylalanine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Phenylalanine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenylalanine or derivatives
- 3-phenylpropanoic-acid
- Phenyl phosphate
- Alpha-amino acid
- L-alpha-amino acid
- Amphetamine or derivatives
- Aryl phosphomonoester
- Aryl phosphate
- Phenoxy compound
- Aralkylamine
- Phosphoric acid ester
- Monocyclic benzene moiety
- Organic phosphoric acid derivative
- Benzenoid
- Amino acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Primary amine
- Hydrocarbon derivative
- Primary aliphatic amine
- Organic oxide
- Carbonyl group
- Organopnictogen compound
- Organic oxygen compound
- Amine
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00kv-9760000000-fac0ad90cc9632ea6e27 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-014i-7490000000-942403f966393fcd98df | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-02td-1190000000-c4bdb7da24563b4177c3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014j-1790000000-55cef97dac3a8d00ae8e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-2900000000-1f6422b48ae626b43485 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9030000000-8821df5eb9bab97574d6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9110000000-52223829a773b1820856 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-0e7b4ff762027c709269 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-9060000000-ccb200a7330d4d29af62 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-a5e502a2627af2048a1f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-a5e502a2627af2048a1f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-02t9-0090000000-813cf20537b120ee03f1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014j-1190000000-30d2bc5ccbdd91a0b720 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-014i-3910000000-6669265710f2d4346cd6 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kataoka H, Nakai K, Makita M: Increase of phosphotyrosine levels in mouse urine and liver during liver regeneration after partial hepatectomy. Biochem Biophys Res Commun. 1994 Jun 15;201(2):909-16. Pubmed: 7516161
- Kataoka, H., Nakai, K., Katagiri, Y., Makita, M. (1993). "Analysis of free and bound O-phosphoamino acids in urine by gas chromatography with flame photometric detection." Biomed Chromatogr 7:184-188. Pubmed: 7693088
- Munoz GE, Arenas-Diaz G, Marshall SH: Exogenously added free phosphotyrosine induces aggregation of circulating platelets in rabbits. Cell Mol Biol (Noisy-le-grand). 1992 Nov;38(7):719-22. Pubmed: 1282059
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|