| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-07-30 14:55:09 -0600 |
|---|
| Update Date | 2015-06-03 17:21:02 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
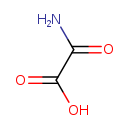
| Name: | Oxamate |
|---|
| Description | Oxamate is an amino-substituted glyoxylic acid derivative. It is believed to be a product of Allantoin degradation in E. coli. Enteric bacteria such as E. coli are able to utilize allantoin as a sole source of nitrogen under anaerobic conditions, but can not utilize it as a sole source of carbon. The first step in allantoin degradation is the opening of the aromatic ring, yielding allantoate, performed by allantoinase. In the next step allantoate is hydrolyzed to S-ureidoglycine by allantoate amidohydrolase. Ureidoglycolate dehydrogenase then oxidizes ureidoglycolate to oxalurate. It is believed oxalurate is converted into oxamate and carbamoyl-phosphate, which can be further metabolized to CO2, ammonia and ATP. |
|---|
| Structure | |
|---|
| Synonyms: | - (aminocarbonyl)-Formate
- (aminocarbonyl)-Formic acid
- (aminocarbonyl)-Oxamate
- (Aminocarbonyl)-oxamic acid
- 2-Oxo-Glycine
- Amino(oxo)acetate
- Amino(oxo)acetic acid
- Amino-Glyoxylate
- Amino-Glyoxylic acid
- Aminooxo-Acetate
- Aminooxo-Acetic acid
- Carbamoyl-Formate
- Carbamoyl-Formic acid
- Iodoxamate meglumine (USAN)
- Iodoxamic acid meglumine (USAN)
- OA (VAN)
- Oxalamate
- Oxalamic acid
- Oxalate monoamide
- Oxalic acid monoamide
- Oxalic monoamide
- Oxamate
- Oxamic acid
- Oxamidate
- Oxamidic acid
|
|---|
| Chemical Formula: | C2H3NO3 |
|---|
| Weight: | Average: 89.0501
Monoisotopic: 89.011292967 |
|---|
| InChI Key: | SOWBFZRMHSNYGE-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C2H3NO3/c3-1(4)2(5)6/h(H2,3,4)(H,5,6) |
|---|
| CAS number: | 471-47-6 |
|---|
| IUPAC Name: | carbamoylformic acid |
|---|
| Traditional IUPAC Name: | oxamic acid |
|---|
| SMILES: | NC(=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as alpha amino acids and derivatives. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon), or a derivative thereof. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Alpha amino acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-amino acid or derivatives
- Primary carboxylic acid amide
- Carboxamide group
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -1 |
|---|
| Melting point: | 210 °C |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | allantoin degradation (anaerobic) | PW002050 |    |
|
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | - allantoin degradation IV (anaerobic) PWY0-41
|
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|