Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramic acid (M2MDB001656)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-07-30 14:55:06 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:21:00 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
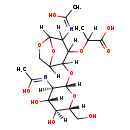
| Name: | N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | N-acetyl-d-glucosamine(anhydrous)n-acetylmuramic acid belongs to the class of Hexoses. These are monosaccharides in which the sugar unit is a hexose. (inferred from compound structure) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C19H30N2O12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 478.4477 Monoisotopic: 478.179874434 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | MWWQKONGFKUAEK-AJSMYUJSSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C19H30N2O12/c1-6(17(27)28)30-16-12(21-8(3)24)18-29-5-10(32-18)15(16)33-19-11(20-7(2)23)14(26)13(25)9(4-22)31-19/h6,9-16,18-19,22,25-26H,4-5H2,1-3H3,(H,20,23)(H,21,24)(H,27,28)/t6?,9-,10?,11-,12?,13-,14-,15?,16?,18?,19+/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2-[(2-{[(2S,3R,4R,5S,6R)-4,5-dihydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-2-yl]oxy}-4-[(1-hydroxyethylidene)amino]-6,8-dioxabicyclo[3.2.1]octan-3-yl)oxy]propanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 2-[(2-{[(2S,3R,4R,5S,6R)-4,5-dihydroxy-3-[(1-hydroxyethylidene)amino]-6-(hydroxymethyl)oxan-2-yl]oxy}-4-[(1-hydroxyethylidene)amino]-6,8-dioxabicyclo[3.2.1]octan-3-yl)oxy]propanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC(OC1C(O[C@@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2N=C(C)O)C2COC(O2)C1N=C(C)O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as n-acyl-alpha-hexosamines. These are carbohydrate derivatives containing a hexose moiety in which the oxygen atom is replaced by an n-acyl group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | N-acyl-alpha-hexosamines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramyl-tetrapeptide + Water > L-Alanine-D-glutamate-meso-2,6-diaminoheptanedioate-D-alanine + N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramic acid N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramyl-tripeptide + Water > L-alanine-D-glutamate-meso-2,6-diaminoheptanedioate + N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramic acid N-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramic acid + Water > N-Acetyl-D-glucosamine + 1,6-Anhydro-N-acetylmuramate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in N-acetylmuramoyl-L-alanine amidase activity
- Specific function:
- Cell-wall hydrolase probably involved in cell-wall hydrolysis, septation or recycling
- Gene Name:
- amiB
- Uniprot ID:
- P26365
- Molecular weight:
- 47985
Reactions
| Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides. |
- General function:
- Involved in N-acetylmuramoyl-L-alanine amidase activity
- Specific function:
- Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides
- Gene Name:
- amiA
- Uniprot ID:
- P36548
- Molecular weight:
- 31412
Reactions
| Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides. |
- General function:
- Involved in N-acetylmuramoyl-L-alanine amidase activity
- Specific function:
- Cell-wall hydrolase probably involved in cell-wall hydrolysis, septation or recycling
- Gene Name:
- amiC
- Uniprot ID:
- P63883
- Molecular weight:
- 45634
Reactions
| Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides. |
- General function:
- Involved in beta-N-acetylhexosaminidase activity
- Specific function:
- Cleaves GlcNAc linked beta-1,4 to MurNAc tripeptides
- Gene Name:
- nagZ
- Uniprot ID:
- P75949
- Molecular weight:
- 37594
Reactions
| Hydrolysis of terminal non-reducing N-acetyl-D-hexosamine residues in N-acetyl-beta-D-hexosaminides. |
- General function:
- Involved in N-acetylmuramoyl-L-alanine amidase activity
- Specific function:
- Involved in both cell wall peptidoglycans recycling and beta-lactamase induction. Specifically cleaves the amide bond between the lactyl group of N-acetylmuramic acid and the alpha- amino group of the L-alanine in degradation products containing an anhydro N-acetylmuramyl moiety
- Gene Name:
- ampD
- Uniprot ID:
- P13016
- Molecular weight:
- 20536
Reactions
| Hydrolyzes the link between N-acetylmuramoyl residues and L-amino acid residues in certain cell-wall glycopeptides. |
Transporters
- General function:
- Involved in transmembrane transport
- Specific function:
- Probably acts as a permease in the beta-lactamase induction system and in peptidoglycan recycling
- Gene Name:
- ampG
- Uniprot ID:
- P0AE16
- Molecular weight:
- 53245
- General function:
- Involved in transporter activity
- Specific function:
- Non-specific porin
- Gene Name:
- ompN
- Uniprot ID:
- P77747
- Molecular weight:
- 41220
- General function:
- Involved in transporter activity
- Specific function:
- Uptake of inorganic phosphate, phosphorylated compounds, and some other negatively charged solutes
- Gene Name:
- phoE
- Uniprot ID:
- P02932
- Molecular weight:
- 38922
- General function:
- Involved in transporter activity
- Specific function:
- OmpF is a porin that forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane. It is also a receptor for the bacteriophage T2
- Gene Name:
- ompF
- Uniprot ID:
- P02931
- Molecular weight:
- 39333
- General function:
- Involved in transporter activity
- Specific function:
- Forms passive diffusion pores which allow small molecular weight hydrophilic materials across the outer membrane
- Gene Name:
- ompC
- Uniprot ID:
- P06996
- Molecular weight:
- 40368