Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Ferrichrome (M2MDB001626)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-07-30 14:54:59 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:20:55 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Ferrichrome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
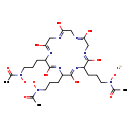
| Description | Ferrichrome is a cyclic hexa-peptide that forms a complex with iron atoms. It is a siderophore composed of three glycine and three modified ornithine residues with hydroxamate groups [-N(OH)C(=O)C-]. The 6 oxygen atoms from the three hydroxamate groups bind Fe(III) in near perfect octahedral coordination. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C27H42FeN9O12 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 740.52 Monoisotopic: 740.230234986 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | GGUNGDGGXMHBMJ-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C27H42N9O12.Fe/c1-16(37)34(46)10-4-7-19-25(43)30-14-23(41)28-13-22(40)29-15-24(42)31-20(8-5-11-35(47)17(2)38)26(44)33-21(27(45)32-19)9-6-12-36(48)18(3)39;/h19-21H,4-15H2,1-3H3,(H,28,41)(H,29,40)(H,30,43)(H,31,42)(H,32,45)(H,33,44);/q-3;+3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 15630-64-5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 1-{3,29-diacetyl-9,12,15,18,21,34-hexahydroxy-2,27,28-trioxa-3,8,11,14,17,20,26,29,33-nonaaza-1-ferratricyclo[17.8.5.2⁷,²²]tetratriaconta-8,11,14,17,20,33-hexaen-26-yl}ethan-1-one | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 1-{3,29-diacetyl-9,12,15,18,21,34-hexahydroxy-2,27,28-trioxa-3,8,11,14,17,20,26,29,33-nonaaza-1-ferratricyclo[17.8.5.2⁷,²²]tetratriaconta-8,11,14,17,20,33-hexaen-26-yl}ethanone | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC(=O)N1CCCC2N=C(O)CN=C(O)CN=C(O)CN=C(O)C3CCCN(O[Fe](O1)ON(CCCC(N=C2O)C(O)=N3)C(C)=O)C(C)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as oligopeptides. These are organic compounds containing a sequence of between three and ten alpha-amino acids joined by peptide bonds. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Oligopeptides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex fhuCDB involved in iron(3+)-hydroxamate import. Responsible for energy coupling to the transport system
- Gene Name:
- fhuC
- Uniprot ID:
- P07821
- Molecular weight:
- 28886
Reactions
| ATP + H(2)O + iron chelate(Out) = ADP + phosphate + iron chelate(In). |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the ABC transporter complex fhuCDB involved in iron(3+)-hydroxamate import. Responsible for the translocation of the substrate across the membrane
- Gene Name:
- fhuB
- Uniprot ID:
- P06972
- Molecular weight:
- 70422
- General function:
- Involved in binding
- Specific function:
- Part of the ABC transporter complex fhuCDB involved in iron(3+)-hydroxamate import. Binds the iron(3+)-hydroxamate complex and transfers it to the membrane-bound permease. Required for the transport of all iron(3+)-hydroxamate siderophores such as ferrichrome, gallichrome, desferrioxamine, coprogen, aerobactin, shizokinen, rhodotorulic acid and the antibiotic albomycin
- Gene Name:
- fhuD
- Uniprot ID:
- P07822
- Molecular weight:
- 32998
Transporters
- General function:
- Involved in nucleotide binding
- Specific function:
- Part of the ABC transporter complex fhuCDB involved in iron(3+)-hydroxamate import. Responsible for energy coupling to the transport system
- Gene Name:
- fhuC
- Uniprot ID:
- P07821
- Molecular weight:
- 28886
Reactions
| ATP + H(2)O + iron chelate(Out) = ADP + phosphate + iron chelate(In). |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the ABC transporter complex fhuCDB involved in iron(3+)-hydroxamate import. Responsible for the translocation of the substrate across the membrane
- Gene Name:
- fhuB
- Uniprot ID:
- P06972
- Molecular weight:
- 70422
- General function:
- Involved in receptor activity
- Specific function:
- This receptor binds the ferrichrome-iron ligand. It interacts with the tonB protein, which is responsible for energy coupling of the ferrichrome-promoted iron transport system. Acts as a receptor for bacteriophage T5 as well as T1, phi80 and colicin M. Binding of T5 triggers the opening of a high conductance ion channel. Can also transport the antibiotic albomycin
- Gene Name:
- fhuA
- Uniprot ID:
- P06971
- Molecular weight:
- 82182
- General function:
- Involved in protein transporter activity
- Specific function:
- Involved in the tonB-dependent energy-dependent transport of various receptor-bound substrates. Protects exbD from proteolytic degradation and functionally stabilizes tonB
- Gene Name:
- exbB
- Uniprot ID:
- P0ABU7
- Molecular weight:
- 26287
- General function:
- Involved in transporter activity
- Specific function:
- Involved in the tonB-dependent energy-dependent transport of various receptor-bound substrates
- Gene Name:
- exbD
- Uniprot ID:
- P0ABV2
- Molecular weight:
- 15527
- General function:
- Involved in iron ion transmembrane transporter activity
- Specific function:
- Interacts with outer membrane receptor proteins that carry out high-affinity binding and energy dependent uptake into the periplasmic space of specific substrates such as cobalamin, and various iron compounds (such as iron dicitrate, enterochelin, aerobactin, etc.). In the absence of tonB these receptors bind their substrates but do not carry out active transport. TonB also interacts with some colicins and is involved in the energy- dependent, irreversible steps of bacteriophages phi 80 and T1 infection. It could act to transduce energy from the cytoplasmic membrane to specific energy-requiring processes in the outer membrane, resulting in the release into the periplasm of ligands bound by these outer membrane proteins
- Gene Name:
- tonB
- Uniprot ID:
- P02929
- Molecular weight:
- 26094
- General function:
- Involved in binding
- Specific function:
- Part of the ABC transporter complex fhuCDB involved in iron(3+)-hydroxamate import. Binds the iron(3+)-hydroxamate complex and transfers it to the membrane-bound permease. Required for the transport of all iron(3+)-hydroxamate siderophores such as ferrichrome, gallichrome, desferrioxamine, coprogen, aerobactin, shizokinen, rhodotorulic acid and the antibiotic albomycin
- Gene Name:
- fhuD
- Uniprot ID:
- P07822
- Molecular weight:
- 32998