| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-07-30 14:54:46 -0600 |
|---|
| Update Date | 2015-06-03 17:20:48 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
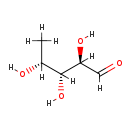
| Name: | 5'-Deoxyribose |
|---|
| Description | 5'-deoxyribose is a deoxy pentose meaning that it is derived from the sugar ribose by loss of an oxygen atom in the 5' position. The more common form of deoxyribose used in DNA is 2'deoxyribose. 5'-deoxyribose can be formed by the breakdown of 5'deoxyadenosine as catalyzed by 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase. |
|---|
| Structure | |
|---|
| Synonyms: | - (2R,3R,4R)-2,3,4-trihydroxypentanal
- (2R,3R,4R)-2,3,4-trihydroxyvaleraldehyde
- 5'-Deoxyribose
- 5-Deoxy-aldehydo-D-ribose
- 5-Deoxy-D-ribose
- 5-Desoxy-D-ribose
- D-Ribo-2,3,4-trihydroxyvaleraldehyde
|
|---|
| Chemical Formula: | C5H10O4 |
|---|
| Weight: | Average: 134.1305
Monoisotopic: 134.057908808 |
|---|
| InChI Key: | WDRISBUVHBMJEF-MROZADKFSA-N |
|---|
| InChI: | InChI=1S/C5H10O4/c1-3(7)5(9)4(8)2-6/h2-5,7-9H,1H3/t3-,4+,5-/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2R,3R,4R)-2,3,4-trihydroxypentanal |
|---|
| Traditional IUPAC Name: | 5'-deoxyribose |
|---|
| SMILES: | [H]O[C@@]([H])(C([H])=O)[C@]([H])(O[H])[C@]([H])(O[H])C([H])([H])[H] |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as monosaccharides. Monosaccharides are compounds containing one carbohydrate unit not glycosidically linked to another such unit, and no set of two or more glycosidically linked carbohydrate units. Monosaccharides have the general formula CnH2nOn. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Monosaccharides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Monosaccharide
- Beta-hydroxy aldehyde
- Alpha-hydroxyaldehyde
- Secondary alcohol
- Polyol
- Organic oxide
- Hydrocarbon derivative
- Short-chain aldehyde
- Carbonyl group
- Aldehyde
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 0 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | Not Available |
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|