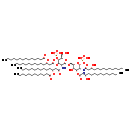| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:58:23 -0600 |
|---|
| Update Date | 2015-06-03 17:20:37 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Lipid A |
|---|
| Description | Lipid A is a lipid that comprises the innermost of the three regions of the lipopolysaccharide (LPS) molecule found in the outer membrane of Gram-negative bacteria such as E. coli. Its hydrophobic nature allows it to anchor the LPS to the outer membrane. Lipid A consists of two glucosamine (carbohydrate/sugar) units with attached acyl chains ("fatty acids"), and normally containing one phosphate group on each carbohydrate. The E. coli lipid A typically has four C14 hydroxy acyl chains attached to the sugars and one C14 and one C12 attached to the beta hydroxy groups. (Wikipedia) |
|---|
| Structure | |
|---|
| Synonyms: | - Diphosphoryl hexaacyl lipid A
- NULL
- Synthetic E. coli lipid A
|
|---|
| Chemical Formula: | C94H178N2O25P2 |
|---|
| Weight: | Average: 1798.365
Monoisotopic: 1797.21939228 |
|---|
| InChI Key: | GZQKNULLWNGMCW-PWQABINMSA-N |
|---|
| InChI: | InChI=1S/C94H178N2O25P2/c1-7-13-19-25-31-37-38-44-50-56-62-68-84(103)115-78(66-60-54-48-42-35-29-23-17-11-5)72-86(105)119-92-88(96-82(101)71-77(65-59-53-47-41-34-28-22-16-10-4)114-83(102)67-61-55-49-43-36-30-24-18-12-6)93(116-79(73-97)90(92)120-122(107,108)109)113-74-80-89(106)91(118-85(104)70-76(99)64-58-52-46-40-33-27-21-15-9-3)87(94(117-80)121-123(110,111)112)95-81(100)69-75(98)63-57-51-45-39-32-26-20-14-8-2/h75-80,87-94,97-99,106H,7-74H2,1-6H3,(H,95,100)(H,96,101)(H2,107,108,109)(H2,110,111,112)/t75-,76-,77-,78-,79-,80-,87-,88-,89-,90-,91-,92-,93-,94-/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | {[(2R,3S,4R,5R,6R)-5-[(3R)-3-(dodecanoyloxy)tetradecanamido]-6-{[(2R,3S,4R,5R,6R)-3-hydroxy-5-[(3R)-3-hydroxytetradecanamido]-4-{[(3R)-3-hydroxytetradecanoyl]oxy}-6-(phosphonooxy)oxan-2-yl]methoxy}-2-(hydroxymethyl)-4-{[(3R)-3-(tetradecanoyloxy)tetradecanoyl]oxy}oxan-3-yl]oxy}phosphonic acid |
|---|
| Traditional IUPAC Name: | lipid A |
|---|
| SMILES: | CCCCCCCCCCCCCC(=O)O[C@H](CCCCCCCCCCC)CC(=O)O[C@@H]1[C@@H](NC(=O)C[C@@H](CCCCCCCCCCC)OC(=O)CCCCCCCCCCC)[C@H](OC[C@H]2O[C@H](OP(O)(O)=O)[C@H](NC(=O)C[C@H](O)CCCCCCCCCCC)[C@@H](OC(=O)C[C@H](O)CCCCCCCCCCC)[C@@H]2O)O[C@H](CO)[C@H]1OP(O)(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as acylaminosugars. These are organic compounds containing a sugar linked to a chain through N-acyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Acylaminosugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Acylaminosugar
- Saccharolipid
- Hexose phosphate
- Disaccharide phosphate
- N-acyl-alpha-hexosamine
- Tetracarboxylic acid or derivatives
- Disaccharide
- Glycosyl compound
- O-glycosyl compound
- Monoalkyl phosphate
- Beta-hydroxy acid
- Fatty acid ester
- Alkyl phosphate
- Fatty acyl
- Phosphoric acid ester
- Oxane
- Organic phosphoric acid derivative
- N-acyl-amine
- Hydroxy acid
- Fatty amide
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxamide group
- Carboxylic acid ester
- Oxacycle
- Organoheterocyclic compound
- Carboxylic acid derivative
- Acetal
- Hydrocarbon derivative
- Primary alcohol
- Alcohol
- Carbonyl group
- Organonitrogen compound
- Organopnictogen compound
- Organic oxide
- Organic nitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -4 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Membrane |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | - Lipopolysaccharide biosynthesis ec00540
|
|---|
| EcoCyc Pathways: | - superpathway of lipopolysaccharide biosynthesis LPSSYN-PWY
|
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0089-0123161900-a0b29cb2a5620669c282 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ff1-1223093400-c7c1cee9a9256bd28b6e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00e9-0242489600-77966b68d48be8286cb4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002b-9510133200-a35ef4a869747eee82f3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9010121101-806a5bc007a2af4acfae | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-0c15069faff9658a6001 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004j-1290020200-6ff62fce7a1fc5f77865 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-4390030000-fc40ff09fa4e7b1694bf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-056v-9640124000-40187fa65042441659ca | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ul1-0001090300-7844b0b3a9cc31e69e69 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0kor-3740180900-796c703855f3e6dcf409 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-4910012000-0ea888969ae27836334e | View in MoNA |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Uniprot Consortium (2012). "Reorganizing the protein space at the Universal Protein Resource (UniProt)." Nucleic Acids Res 40:D71-D75. Pubmed: 22102590
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|