| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:54:10 -0600 |
|---|
| Update Date | 2015-06-03 17:20:24 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Cinnavalininate |
|---|
| Description | Cinnavalininate is an intermediate in the tryptophan metabolic pathway [Kegg: C05640]. It is generated from 3-hydroxyanthranilate via the enzyme catalase (EC:1.11.1.6). |
|---|
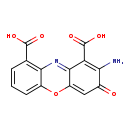
| Structure | |
|---|
| Synonyms: | - 2-Amino-3-oxo-3H-phenoxazin-1,9-dicarboxylate
- 2-Amino-3-oxo-3H-phenoxazin-1,9-dicarboxylic acid
- 2-Amino-3H-phenoxazin-one-1,9-dicarboxylate
- 2-Amino-3H-phenoxazin-one-1,9-dicarboxylic acid
- Cinnabarinate
- Cinnabarinic acid
- Cinnavalininic acid
|
|---|
| Chemical Formula: | C14H8N2O6 |
|---|
| Weight: | Average: 300.2231
Monoisotopic: 300.038235998 |
|---|
| InChI Key: | FSBKJYLVDRVPTK-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C14H8N2O6/c15-10-6(17)4-8-12(9(10)14(20)21)16-11-5(13(18)19)2-1-3-7(11)22-8/h1-4H,15H2,(H,18,19)(H,20,21) |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | 2-amino-3-oxo-3H-phenoxazine-1,9-dicarboxylic acid |
|---|
| Traditional IUPAC Name: | cinnabarinic acid |
|---|
| SMILES: | NC1=C(C(O)=O)C2=NC3=C(C=CC=C3OC2=CC1=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phenoxazines. These are polycyclic aromatic compounds containing a phenoxazine moiety, which is a linear tricyclic system that consists of a two benzene rings joined by a 1,4-oxazine ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Benzoxazines |
|---|
| Sub Class | Phenoxazines |
|---|
| Direct Parent | Phenoxazines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenoxazine
- Dicarboxylic acid or derivatives
- Benzenoid
- Heteroaromatic compound
- Vinylogous amide
- Cyclic ketone
- Amino acid
- Amino acid or derivatives
- Carboxylic acid derivative
- Carboxylic acid
- Oxacycle
- Azacycle
- Primary amine
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Amine
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0kal-1090000000-a9b3f04e0154e56a34ac | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-05fr-7019200000-9d027bf9474c784d794f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0093000000-6eec5362c826437aa9f8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a59-0090000000-ea4d6ec354a7ad615bcd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a5i-2090000000-a1cd4541b5bfb101f231 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0090000000-7b8418c9fe543cb0ddfc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0bt9-0090000000-3aa6f5b7cbca4210c35f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0bt9-0390000000-10af92579df64b8720cb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0090000000-fb1be335f42676557e20 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0bti-0090000000-54b079267e6329486c07 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03fr-0190000000-40328130c8a319d1d980 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0090000000-24fd7e79c1bc515afade | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0159-0090000000-dadf0f0bb8448267e058 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0090000000-362869951e6e3adac9c6 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | HMDB04078 | | Pubchem Compound ID | 114918 | | Kegg ID | C05640 | | ChemSpider ID | 102864 | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|