Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
L-Seryl-AMP (M2MDB001407)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:53:09 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:20:22 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
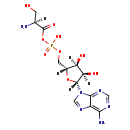
| Name: | L-Seryl-AMP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | L-Seryl-AMP is an enzyme-bound intermediate found in the protein EntF. EntF is the enzyme responsible for serine activation during the biosynthesis of enterobactin (a cyclic trimer of N-dihydroxybenzoyl serine) in Escherichia coli. Serine adenylate is also an intermediate in the charging of Seryl-tRNA-synthetase. In particular, Mg.ATP and serine react to form seryl-adenylate on the protein. The serine is subsequently transferred to the 3'-end of the tRNA. Seryl-tRNA synthetase is also capable of synthesizing diadenosine tetraphosphate (Ap4A) from the enzyme-bound seryl-adenylate intermediate and a second molecule of ATP. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C13H19N6O9P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 434.2985 Monoisotopic: 434.095112748 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | UVSYURUCZPPUQD-MACXSXHHSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C13H19N6O9P/c14-5(1-20)13(23)28-29(24,25)26-2-6-8(21)9(22)12(27-6)19-4-18-7-10(15)16-3-17-11(7)19/h3-6,8-9,12,20-22H,1-2,14H2,(H,24,25)(H2,15,16,17)/t5-,6+,8+,9+,12+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 52435-67-3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(2S)-2-amino-3-hydroxypropanoyl]oxy}({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy})phosphinic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | L-seryl-AMP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@](N)(CO)C(=O)OP(O)(=O)OC[C@@]1([H])O[C@@]([H])(N2C=NC3=C(N)N=CN=C23)[C@]([H])(O)[C@]1([H])O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as 5'-acylphosphoadenosines. These are ribonucleoside derivatives containing an adenoside moiety, where the phosphate group is acylated. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Purine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine ribonucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | 5'-acylphosphoadenosines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 3 (2,3-Dihydroxybenzoyl)adenylic acid + 3 L-Seryl-AMP >6 Adenosine monophosphate + Enterochelin +9 Hydrogen ion Adenosine triphosphate + Hydrogen ion + L-Serine <> Pyrophosphate + L-Seryl-AMP Adenosine triphosphate + L-Serine <> Pyrophosphate + L-Seryl-AMP (2,3-Dihydroxybenzoyl)adenylic acid + L-Seryl-AMP <> Hydrogen ion + Enterochelin + Adenosine monophosphate Adenosine triphosphate + L-Serine > Pyrophosphate + L-Seryl-AMP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in ligase activity
- Specific function:
- Activates the carboxylate group of L-serine via ATP- dependent PPi exchange reactions to the aminoacyladenylate, preparing that molecule for the final stages of enterobactin synthesis. Holo-entF acts as the catalyst for the formation of the three amide and three ester bonds present in the cyclic (2,3- dihydroxybenzoyl)serine trimer enterobactin, using seryladenylate and acyl-holo-entB (acylated with 2,3-dihydroxybenzoate by entE)
- Gene Name:
- entF
- Uniprot ID:
- P11454
- Molecular weight:
- 141990
Reactions
| ATP + L-serine = diphosphate + L-serine-adenylate. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Catalyzes the transfer of the 4'-phosphopantetheine moiety from coenzyme A to apo-domains of both entB (an ArCP domain) and entF (a PCP domain). Plays an essential role in the assembly of the enterobactin
- Gene Name:
- entD
- Uniprot ID:
- P19925
- Molecular weight:
- 23604
Reactions
| CoA + apo-EntB/F = adenosine 3',5'-bisphosphate + holo-EntB/F. |