| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:52:44 -0600 |
|---|
| Update Date | 2015-06-03 17:20:22 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
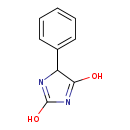
| Name: | Phenylhydantoin |
|---|
| Description | Phenylhydantoin is a cyclic ureide and it is believed that E. coli may use these compounds as nutrient sources. The gene hyuA codes for d-stereospecific phenylhydantoinase (465 amino acids) which is a homotetramer. This enzyme belongs to the cyclic amidohydrolase superfamily. E. coli D-phenylhydantoinase exhibits a distinct activity toward phenylhydantoin with an aromatic side chain at the 5' position but does not readily hydrolyze the simple cyclic ureides. D-phenylhydantoinase shares a significant homology (>45%) with those of allantoinase and dihydropyrimidinase. |
|---|
| Structure | |
|---|
| Synonyms: | - 5-Phenyl-(.+-.)-2,4-Imidazolidinedione
- 5-Phenyl-2,4-Imidazolidinedione
- 5-Phenyl-Hydantoin
- 5-Phenylhydantoin
- Hydantoin, 5-phenyl- (8CI)
|
|---|
| Chemical Formula: | C9H8N2O2 |
|---|
| Weight: | Average: 176.172
Monoisotopic: 176.05857751 |
|---|
| InChI Key: | NXQJDVBMMRCKQG-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C9H8N2O2/c12-8-7(10-9(13)11-8)6-4-2-1-3-5-6/h1-5,7H,(H2,10,11,12,13) |
|---|
| CAS number: | 89-24-7 |
|---|
| IUPAC Name: | 4-phenyl-4H-imidazole-2,5-diol |
|---|
| Traditional IUPAC Name: | 5-phenyl-5H-imidazole-2,4-diol |
|---|
| SMILES: | OC1=NC(C(O)=N1)C1=CC=CC=C1 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phenylhydantoins. These are heterocyclic aromatic compounds containing an imiazolidinedione moiety substituted by a phenyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Azolidines |
|---|
| Sub Class | Imidazolidines |
|---|
| Direct Parent | Phenylhydantoins |
|---|
| Alternative Parents | |
|---|
| Substituents | - 5-phenylhydantoin
- Phenylimidazolidine
- Alpha-amino acid or derivatives
- 5-monosubstituted hydantoin
- N-acyl urea
- Ureide
- Monocyclic benzene moiety
- Benzenoid
- Dicarboximide
- Urea
- Carbonic acid derivative
- Carboxylic acid derivative
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic nitrogen compound
- Carbonyl group
- Organic oxygen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | | Collection of Reactions without pathways | PW001891 |    |
|
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-001i-2900000000-29031da4f3df492b09a7 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0900000000-b8ee69b6ad9468557700 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0zi0-0900000000-2608f617a5a27f1e6151 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0zfr-6900000000-0c826d2a1ec8a246a52d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-2900000000-2fd3b420a7b79ffe7263 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-002f-9700000000-5ba49553bdd1e846ac2d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0f6x-9500000000-396218cd005d46fe3c61 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-004i-0900000000-72b2beb77ff784d05490 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ugi-2900000000-0c30a9d25db568f80182 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-4900000000-accbe98700d8ea46df6e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-ca891ec865f0b18f774e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-054o-9700000000-2b7dcd52fe2a35e704b3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9300000000-2eec2b1f43ac2ef28d16 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|