| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:50:47 -0600 |
|---|
| Update Date | 2015-06-03 17:20:18 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | GMP-Lysine |
|---|
| Description | GMP-lysine is a member of the chemical class known as Purine Ribonucleoside Monophosphates. These are nucleotides consisting of a purine base linked to a ribose to which one monophosphate group is attached. GMP-lysine is a nucleotidyl-modified amino acid. The hinT gene product is capable of cleaving lysine from GMP and AMP. Though specific protein targets remain to be identified, it is has been hypothesized that Hint hydrolases may reverse nucleotidylylated protein modifications of lysine. In E. coli, the Hint-homologous hinT gene is required for resistance to elevated levels of certain salts. |
|---|
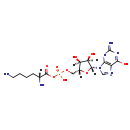
| Structure | |
|---|
| Synonyms: | |
|---|
| Chemical Formula: | C16H27N7O9P |
|---|
| Weight: | Average: 492.4008
Monoisotopic: 492.160787009 |
|---|
| InChI Key: | XRYPUFFHVWYVTB-BAYCTPFLSA-P |
|---|
| InChI: | InChI=1S/C16H25N7O9P/c17-4-2-1-3-7(18)15(27)32-33(28,29)30-5-8-10(24)11(25)14(31-8)23-6-20-9-12(23)21-16(19)22-13(9)26/h6-8,10-11,14,24-25H,1-5,17-18H2,(H3-,19,21,22,26,28,29)/p+2/t7-,8-,10-,11-,14-/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | 9-[(2R,3R,4S,5R)-5-{[({[(2R)-2,6-diazaniumylhexanoyl]oxy}(hydroxy)phosphoryl)oxy]methyl}-3,4-dihydroxyoxolan-2-yl]-6-hydroxy-2-imino-3,9-dihydro-2H-7λ⁵-purin-7-ylium-3-id-7-yl |
|---|
| Traditional IUPAC Name: | 9-[(2R,3R,4S,5R)-5-[({[(2R)-2,6-diammoniohexanoyl]oxy(hydroxy)phosphoryl}oxy)methyl]-3,4-dihydroxyoxolan-2-yl]-6-hydroxy-2-imino-3H-7λ⁵-purin-7-ylium-3-id-7-yl |
|---|
| SMILES: | [H][C@@]([NH3+])(CCCC[NH3+])C(=O)OP(O)(=O)OC[C@@]1([H])O[C@@]([H])(N2C=[N+]C3=C2[N-]C(=N)N=C3O)[C@]([H])(O)[C@]1([H])O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine ribonucleoside monophosphates. These are nucleotides consisting of a purine base linked to a ribose to which one monophosphate group is attached. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine ribonucleotides |
|---|
| Direct Parent | Purine ribonucleoside monophosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine ribonucleoside monophosphate
- Pentose-5-phosphate
- Pentose phosphate
- N-glycosyl compound
- Glycosyl compound
- Pentose monosaccharide
- Monosaccharide phosphate
- Alpha-amino acid or derivatives
- Purine
- Imidazopyrimidine
- Hydroxypyrimidine
- Monoalkyl phosphate
- Acyl phosphate
- Alkyl phosphate
- Pyrimidine
- Phosphoric acid ester
- Organic phosphoric acid derivative
- N-substituted imidazole
- Monosaccharide
- Heteroaromatic compound
- Tetrahydrofuran
- Imidazole
- Azole
- Secondary alcohol
- Carboxylic acid salt
- Amino acid or derivatives
- 1,2-diol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organic salt
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Amine
- Alcohol
- Organic cation
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|