| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:48:41 -0600 |
|---|
| Update Date | 2015-06-03 17:20:08 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | 1,6-Anhydro-N-acetylmuramate |
|---|
| Description | 1,6-anhMurNAC is an Amino Sugar. These are compounds having one alcoholic hydroxy group replaced by an amino group; systematically known as x-amino-x-deoxymonosaccharides. 1,6-anhydro-N-acetylmuramic acid (anhMurNAc) is one of the components of murein that is recycled inside the cell. Although exogenously provided anhMurNAc can be taken up by E. coli, it can not serve as the sole source of carbon for growth. |
|---|
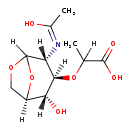
| Structure | |
|---|
| Synonyms: | - 1,6-anhMurNAc
- 1,6-Anhydro-N-acetylmuramate
- 1,6-Anhydro-N-acetylmuramic acid
|
|---|
| Chemical Formula: | C11H17NO7 |
|---|
| Weight: | Average: 275.2552
Monoisotopic: 275.100501903 |
|---|
| InChI Key: | ZFEGYUMHFZOYIY-NFSFVEDMSA-N |
|---|
| InChI: | InChI=1S/C11H17NO7/c1-4(10(15)16)18-9-7(12-5(2)13)11-17-3-6(19-11)8(9)14/h4,6-9,11,14H,3H2,1-2H3,(H,12,13)(H,15,16)/t4?,6-,7-,8-,9-,11?/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | 2-{[(1R,2S,3R,4R)-2-hydroxy-4-[(1-hydroxyethylidene)amino]-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy}propanoic acid |
|---|
| Traditional IUPAC Name: | 2-{[(1R,2S,3R,4R)-2-hydroxy-4-[(1-hydroxyethylidene)amino]-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy}propanoic acid |
|---|
| SMILES: | [H]C(C)(O[C@@]1([H])[C@]([H])(O)[C@@]2([H])COC([H])(O2)[C@]1([H])N=C(C)O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as oxepanes. Oxepanes are compounds containing an oxepane ring, which is a seven-member saturated aliphatic heterocycle with one oxygen and six carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Oxepanes |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Oxepanes |
|---|
| Alternative Parents | |
|---|
| Substituents | - Oxepane
- Oxane
- Monosaccharide
- Meta-dioxolane
- Secondary alcohol
- Oxacycle
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Monocarboxylic acid or derivatives
- Ether
- Dialkyl ether
- Carboxylic acid
- Carboxylic acid derivative
- Carboximidic acid derivative
- Carboximidic acid
- Acetal
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Alcohol
- Aliphatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aliphatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | 1,6-Anhydrous-N-Acetylmuramyl-tetrapeptide + Water > L-Alanine-D-glutamate-meso-2,6-diaminoheptanedioate-D-alanine + 1,6-Anhydro-N-acetylmuramate1,6-Anhydrous-N-Acetylmuramyl-tripeptide + Water > L-alanine-D-glutamate-meso-2,6-diaminoheptanedioate + 1,6-Anhydro-N-acetylmuramateN-Acetyl-D-glucosamine(anhydrous)N-Acetylmuramic acid + Water > N-Acetyl-D-glucosamine + 1,6-Anhydro-N-acetylmuramate1,6-Anhydro-N-acetylmuramate + Adenosine triphosphate + Water > N-Acetylmuramic acid 6-phosphate + ADP + Hydrogen ion<i>N</i>-acetyl-β-D-glucosamine(anhydrous)-<i>N</i>-acetylmuramate + Water N-Acetyl-D-glucosamine + 1,6-Anhydro-N-acetylmuramateAdenosine triphosphate + Water + 1,6-Anhydro-N-acetylmuramate <> Adenosine diphosphate + MurNAc-6-P + ADP |
|---|
| SMPDB Pathways: | | Amino sugar and nucleotide sugar metabolism III | PW000895 |    |
|
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | - 1,6-anhydro-N-acetylmuramic acid recycling PWY0-1261
|
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | 40666 | | HMDB ID | Not Available | | Pubchem Compound ID | 25201515 | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | CPD0-882 | | EcoCyc ID | CPD0-882 |
|
|---|