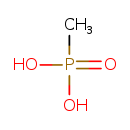| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:39:25 -0600 |
|---|
| Update Date | 2015-06-05 00:35:31 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Methylphosphonate |
|---|
| Description | Methylphosphonate is a member of the chemical class known as Organic Phosphonic Acids and Derivatives. These are organic compounds containing phosphonic acid or a derivative thereof. . Phosphonates (Pn) are a large class of organophosphorus molecules that have direct carbon-phosphorus (C-P) bonds in place of the carbon-oxygen-phosphorus ester bond. In bacteria two pathways exist for Pn breakdown for use as a P source: the phosphonatase and C-P lyase pathways. These pathways differ both in regard to their substrate specificity and their cleavage mechanism. The phosphonatase pathway acts on the natural Pn alpha-aminoethylphosphonate (AEPn). In a two-step process it leads to cleavage of the C-P bond by a hydrolysis reaction requiring an adjacent carbonyl group. In contrast the C-P lyase pathway has a broad substrate specificity. It leads to cleavage of substituted Pn (such as AEPn) as well as unsubstituted Pn by a mechanism involving redox or radical chemistry. Due to its broad substrate specificity, the C-P lyase pathway is generally thought to be responsible for the breakdown of Pn herbicides (such as glyphosate) by bacteria. |
|---|
| Structure | |
|---|
| Synonyms: | - GB
- M-Carboxyphenyl phenylacetamidomethylphosphonate
- M-Carboxyphenyl phenylacetamidomethylphosphonic acid
- MePn
- Methanephosphonate
- Methanephosphonic acid
- Methyl phosphonate
- Methyl phosphonic acid
- Methyl-Phosphonate
- Methyl-Phosphonic acid
- Methylphosphonic acid
- Phosphonomethyl group
- PPM
- VXA
|
|---|
| Chemical Formula: | CH5O3P |
|---|
| Weight: | Average: 96.0224
Monoisotopic: 95.997630538 |
|---|
| InChI Key: | YACKEPLHDIMKIO-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/CH5O3P/c1-5(2,3)4/h1H3,(H2,2,3,4) |
|---|
| CAS number: | 993-13-5 |
|---|
| IUPAC Name: | methylphosphonic acid |
|---|
| Traditional IUPAC Name: | methylphosphonic acid |
|---|
| SMILES: | CP(O)(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as organic phosphonic acids. These are organic compounds containing phosphonic acid. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Organic phosphonic acids and derivatives |
|---|
| Sub Class | Organic phosphonic acids |
|---|
| Direct Parent | Organic phosphonic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Organophosphonic acid
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Organophosphorus compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -1 |
|---|
| Melting point: | 108.5 °C |
|---|
| Experimental Properties: | | Property | Value | Source |
|---|
| Water Solubility: | 20 mg/mL [ROSENBLATT,DH et al. (1975)] | PhysProp |
|
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | - Phosphonate and phosphinate metabolism ec00440
|
|---|
| EcoCyc Pathways: | |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-953ee27b6d4d39a23d75 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-e9ec8666066269105129 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00ls-9000000000-cfc27ecfe46b6bae99b8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-548aafc3bd5ffd117ef4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-794c4f4a660b80340d80 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-41de8276110f43fd3d44 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-27d68e801482e0c96fc7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0032-9000000000-fae35420c79329557d65 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01t9-9000000000-9e51f8409702ef54b755 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-26a762900090ff5befc9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-fcafc7ee01c9b99fcf8f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-1fff7b868f217d05497e | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|