| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:39:19 -0600 |
|---|
| Update Date | 2015-06-03 17:19:47 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | ppGp |
|---|
| Description | Ppgp belongs to the class of Purine Ribonucleoside 3',5'-Bisphosphates. These are purine ribobucleotides with one phosphate group attached to 3' and 5' hydroxyl groups of the ribose moiety. (inferred from compound structure)<br/><br/>Unusual guanosine nucleotides guanosine 5'-diphosphate 3'-diphosphate (ppGpp, also known as MSI) and guanosine 5'-diphosphate 3'-monophosphate (ppGp, also known as MSIII) accumulate to high concentrations in wild-type cells of Escherichia coli during amino acid starvation. (PMID 6117328) |
|---|
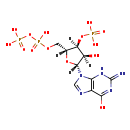
| Structure | |
|---|
| Synonyms: | - Guanosine 5'-diphosphate-3'-monophosphate
- Guanosine 5'-diphosphoric acid-3'-monophosphoric acid
- GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE
- GUANOSINE-3'-monophosphoric acid-5'-diphosphoric acid
- MsIII
|
|---|
| Chemical Formula: | C10H16N5O14P3 |
|---|
| Weight: | Average: 523.1804
Monoisotopic: 522.990659781 |
|---|
| InChI Key: | HEYSFDAMRDTCJM-UUOKFMHZSA-N |
|---|
| InChI: | InChI=1S/C10H16N5O14P3/c11-10-13-7-4(8(17)14-10)12-2-15(7)9-5(16)6(28-30(18,19)20)3(27-9)1-26-32(24,25)29-31(21,22)23/h2-3,5-6,9,16H,1H2,(H,24,25)(H2,18,19,20)(H2,21,22,23)(H3,11,13,14,17)/t3-,5-,6-,9-/m1/s1 |
|---|
| CAS number: | 58902-76-4 |
|---|
| IUPAC Name: | {[(2R,3S,4R,5R)-4-hydroxy-2-({[hydroxy(phosphonooxy)phosphoryl]oxy}methyl)-5-(6-hydroxy-2-imino-3,9-dihydro-2H-purin-9-yl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional IUPAC Name: | [(2R,3S,4R,5R)-4-hydroxy-2-({[hydroxy(phosphonooxy)phosphoryl]oxy}methyl)-5-(6-hydroxy-2-imino-3H-purin-9-yl)oxolan-3-yl]oxyphosphonic acid |
|---|
| SMILES: | [H][C@]1(COP(O)(=O)OP(O)(O)=O)O[C@@]([H])(N2C=NC3=C2NC(=N)N=C3O)[C@]([H])(O)[C@]1([H])OP(O)(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine ribonucleotides |
|---|
| Direct Parent | Purine ribonucleoside diphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose-5-phosphate
- Pentose phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-oxopurine
- Pentose monosaccharide
- Hypoxanthine
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Purine
- Pyrimidone
- Aminopyrimidine
- Monoalkyl phosphate
- Pyrimidine
- Phosphoric acid ester
- Organic phosphoric acid derivative
- N-substituted imidazole
- Monosaccharide
- Alkyl phosphate
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Tetrahydrofuran
- Secondary alcohol
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Organic oxygen compound
- Amine
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Primary amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -4 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | | Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | 188347 | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | CPD0-1055 | | EcoCyc ID | CPD0-1055 | | Ligand Expo | G3D |
|
|---|