| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:36:46 -0600 |
|---|
| Update Date | 2015-09-16 14:50:11 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
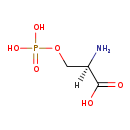
| Name: | O-Phospho-D-serine |
|---|
| Description | O-phospho-D-serine is a phosphoric ester of serine and phosphoric acid. Phosphorylated serine residues are often referred to as phosphoserine. Phosphoserine is a component of many proteins. The phosphorylation of the alcohol functional group in serine to produce phosphoserine is catalyzed by various types of kinases. Free phosphoserine may be found after proteins have been proteolyzed. Phosphoserine phosphatase is able to dephosphorylate phosphoserince (both L and D forms).
|
|---|
| Structure | |
|---|
| Synonyms: | - (2S)-2-amino-3-phosphonooxypropanoate
- (2S)-2-amino-3-phosphonooxypropanoic acid
- 2-Amino-3-(phosphonooxy)propanoate
- 2-Amino-3-(phosphonooxy)propanoic acid
- 3-P-Serine
- 3-Phospho-1-serine
- 3-Phospho-L-serine
- 3-Phospho-serine
- 3-Phosphoserine
- D -2-Amino-3-hydroxypropanoate 3-phosphate
- D -2-Amino-3-hydroxypropanoic acid 3-phosphate
- D -2-amino-3-Hydroxypropanoic acid 3-phosphoric acid
- D-O-Phosphoserine
- DL-2-Amino-3-hydroxypropanoate 3-phosphate
- DL-2-Amino-3-hydroxypropanoic acid 3-phosphate
- DL-2-amino-3-Hydroxypropanoic acid 3-phosphoric acid
- DL-O-phosphorylserine
- DL-O-phosphoserine
- DL-O-serine phosphate
- DL-O-Serine phosphoric acid
- DL-phosphoserine
- DL-serine dihydrogen phosphate
- DL-Serine dihydrogen phosphoric acid
- DL-Serine monophosphate
- DL-serine monophosphorate
- DL-serine monophosphoric acid
- DL-serine, dihydrogen phosphate (ester)
- DL-Serine, dihydrogen phosphoric acid (ester)
- DL-sop
- Energoserina
- L-Serine, dihydrogen phosphate
- L-Serine, dihydrogen phosphoric acid
- O-Phospho-DL-serine
- O-Phospho-L-serine
- O-Phosphono-Serine
- O-Phosphonoserine
- O-Phosphoserine
- P-Ser
- P-Serine
- Phosphonoserine
- Phosphorylserine
- Phosphoserine
- Serine phosphate
- Serine phosphoric acid
- Serine, dihydrogen phosphate (ester) (9CI)
- Serine, dihydrogen phosphate (ester), DL- (8CI)
- Serine, dihydrogen phosphoric acid (ester) (9ci)
- Serine, dihydrogen phosphoric acid (ester), DL- (8ci)
- Serine-3-p
- Serine-3-phosphate
- Serine-3-phosphoric acid
- Serophen
- Seryl phosphate
- Seryl phosphoric acid
- [1-14c] Serine Dihydrogen Phosphate (ester)
- [1-14C] Serine dihydrogen phosphoric acid (ester)
|
|---|
| Chemical Formula: | C3H8NO6P |
|---|
| Weight: | Average: 185.0725
Monoisotopic: 185.008923505 |
|---|
| InChI Key: | BZQFBWGGLXLEPQ-UWTATZPHSA-N |
|---|
| InChI: | InChI=1S/C3H8NO6P/c4-2(3(5)6)1-10-11(7,8)9/h2H,1,4H2,(H,5,6)(H2,7,8,9)/t2-/m1/s1 |
|---|
| CAS number: | 17885-08-4 |
|---|
| IUPAC Name: | (2R)-2-amino-3-(phosphonooxy)propanoic acid |
|---|
| Traditional IUPAC Name: | O-phospho-D-serine |
|---|
| SMILES: | [H][C@@](N)(COP(O)(O)=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as d-alpha-amino acids. These are alpha amino acids which have the D-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | D-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - D-alpha-amino acid
- Phosphoethanolamine
- Monoalkyl phosphate
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Primary amine
- Organopnictogen compound
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Amine
- Primary aliphatic amine
- Carbonyl group
- Organic oxide
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -2 |
|---|
| Melting point: | 228 °C |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | - Glycine, serine and threonine metabolism ec00260
- Metabolic pathways eco01100
- Methane metabolism ec00680
- Microbial metabolism in diverse environments ec01120
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000l-8900000000-fa80206f357e691b1927 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9300000000-db6b02072c0568e8e773 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-b3fe18e3e231ec92a4b7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-003s-8900000000-1624d6cd2551f1de721b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9100000000-6eb41a3603612c36a612 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-cb44ff87fe2054851088 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0007-5900000000-0165c58c1b2c713290ed | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0005-9300000000-8c803e99e72e9cb59913 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000x-9000000000-6ed59f8b5f83d923a8b4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002b-9000000000-68e2a2413db514aab39e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-a5e502a2627af2048a1f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-a5e502a2627af2048a1f | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|