| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:34:40 -0600 |
|---|
| Update Date | 2015-06-03 17:19:36 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | 5-Methylaminomethyl-2-thiouridine |
|---|
| Description | 5-methylaminomethyl-2-thiouridine is a member of the chemical class known as Pyrimidine Nucleosides and Analogues. These are compounds comprising a pyrimidine base attached to a sugar. 5-methylaminomethyl-2-thiouridine is invovled in Selenouridine biosynthesis (tRNA). The selenium-modified nucleoside is 5-methyl-aminomethyl-2-selenouridine (mnm5Se2U), which is the chemical analog of 5-methylaminomethyl-2-thiouridine, a previously identified minor base of E. coli. tRNA2Glu. (PMID 6227514) |
|---|
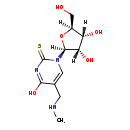
| Structure | |
|---|
| Synonyms: | - 2-Thio-5-methylaminomethyluridine
- 5-((Methylamino)methyl)-2-thio-Uridine
|
|---|
| Chemical Formula: | C11H17N3O5S |
|---|
| Weight: | Average: 303.335
Monoisotopic: 303.088891359 |
|---|
| InChI Key: | HXVKEKIORVUWDR-GHCJXIJMSA-N |
|---|
| InChI: | InChI=1S/C11H17N3O5S/c1-12-2-5-3-14(11(20)13-9(5)18)10-8(17)7(16)6(4-15)19-10/h3,6-8,10,12,15-17H,2,4H2,1H3,(H,13,18,20)/t6-,7-,8-,10-/m0/s1 |
|---|
| CAS number: | 32860-54-1 |
|---|
| IUPAC Name: | 1-[(2S,3S,4R,5S)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-4-hydroxy-5-[(methylamino)methyl]-1,2-dihydropyrimidine-2-thione |
|---|
| Traditional IUPAC Name: | 1-[(2S,3S,4R,5S)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-4-hydroxy-5-[(methylamino)methyl]pyrimidine-2-thione |
|---|
| SMILES: | [H][C@@]1(CO)O[C@]([H])(N2C=C(CNC)C(O)=NC2=S)[C@@]([H])(O)[C@@]1([H])O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine nucleosides. Pyrimidine nucleosides are compounds comprising a pyrimidine base attached to a ribosyl or deoxyribosyl moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleosides |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Pyrimidine nucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine nucleoside
- N-glycosyl compound
- Glycosyl compound
- Pentose monosaccharide
- 2-thiopyrimidine
- Thiopyrimidine
- Aralkylamine
- Pyrimidone
- Pyrimidinethione
- Pyrimidine
- Monosaccharide
- Hydropyrimidine
- Heteroaromatic compound
- Vinylogous amide
- Tetrahydrofuran
- Thiourea
- Secondary alcohol
- Lactam
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Secondary amine
- Secondary aliphatic amine
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Amine
- Alcohol
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | 1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Stadtman, T. C. (1983). "New biologic functions--selenium-dependent nucleic acids and proteins." Fundam Appl Toxicol 3:420-423. Pubmed: 6227514
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|