Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Trans-2,3-Dihydroxycinnamate (M2MDB001043)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:32:38 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-09-17 16:24:21 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Trans-2,3-Dihydroxycinnamate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Trans-2,3-dihydroxycinnamate is a member of the chemical class known as Hydroxycinnamic Acid Derivatives. These are compounds containing an cinnamic acid derivative where the benzene ring is hydroxylated. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
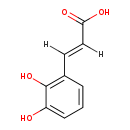
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C9H8O4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 180.1574 Monoisotopic: 180.042258744 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | SIUKXCMDYPYCLH-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C9H8O4/c10-7-3-1-2-6(9(7)13)4-5-8(11)12/h1-5,10,13H,(H,11,12) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 31082-90-3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | trans-2,3-dihydroxycinnamate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC(=O)C=CC1=C(O)C(O)=CC=C1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as hydroxycinnamic acids. Hydroxycinnamic acids are compounds containing an cinnamic acid where the benzene ring is hydroxylated. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Phenylpropanoids and polyketides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Cinnamic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Hydroxycinnamic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Hydroxycinnamic acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic homomonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 3-Hydroxycinnamic acid + Hydrogen ion + NADH + Oxygen > Trans-2,3-Dihydroxycinnamate + Water + NAD Trans-2,3-Dihydroxycinnamate + Oxygen > Hydrogen ion + 2-Hydroxy-6-ketononatrienedioate cis-3-(3-Carboxyethenyl)-3,5-cyclohexadiene-1,2-diol + NAD > Trans-2,3-Dihydroxycinnamate + Hydrogen ion + NADH cis-3-(3-Carboxyethenyl)-3,5-cyclohexadiene-1,2-diol + NAD <> Trans-2,3-Dihydroxycinnamate + NADH + Hydrogen ion 3-Hydroxycinnamic acid + Oxygen + NADH + Hydrogen ion <> Trans-2,3-Dihydroxycinnamate + Water + NAD Trans-2,3-Dihydroxycinnamate + Oxygen <> 2-Hydroxy-6-ketononatrienedioate cis-3-(3-Carboxyethenyl)-3,5-cyclohexadiene-1,2-diol + NAD > Trans-2,3-Dihydroxycinnamate + NADH trans-Cinnamic acid + NADH + Oxygen > Trans-2,3-Dihydroxycinnamate + NAD 3-Hydroxycinnamic acid + NADH + Oxygen > Trans-2,3-Dihydroxycinnamate + Water + NAD Trans-2,3-Dihydroxycinnamate + Oxygen > 2-Hydroxy-6-ketononatrienedioate 3-(3-Hydroxyphenyl)propanoic acid + NADH + Hydrogen ion + Oxygen + 3-Hydroxycinnamic acid <> 3-(2,3-Dihydroxyphenyl)propionic acid + Water + NAD + Trans-2,3-Dihydroxycinnamate 3-(2,3-Dihydroxyphenyl)propionic acid + Oxygen + Trans-2,3-Dihydroxycinnamate <> 2-Hydroxy-6-ketononadienedicarboxylate + 2-Hydroxy-6-ketononatrienedioate NADH + Hydrogen ion + Oxygen + Hydrocinnamic acid <> cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD + Trans-2,3-Dihydroxycinnamate cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD + cis-3-(3-Carboxyethenyl)-3,5-cyclohexadiene-1,2-diol <> 3-(2,3-Dihydroxyphenyl)propionic acid + NADH + Hydrogen ion + Trans-2,3-Dihydroxycinnamate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in iron ion binding
- Specific function:
- Part of the multicomponent 3-phenylpropionate dioxygenase. Converts 3-phenylpropionic acid (PP) and cinnamic acid (CI) into 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol), respectively
- Gene Name:
- hcaE
- Uniprot ID:
- P0ABR5
- Molecular weight:
- 51109
Reactions
| 3-phenylpropanoate + NADH + O(2) = 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD(+). |
| (2E)-3-phenylprop-2-enoate + NADH + O(2) = (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate + NAD(+). |
- General function:
- Involved in iron ion binding
- Specific function:
- Catalyzes the non-heme iron(II)-dependent oxidative cleavage of 2,3-dihydroxyphenylpropionic acid and 2,3- dihydroxicinnamic acid into 2-hydroxy-6-ketononadienedioate and 2- hydroxy-6-ketononatrienedioate, respectively
- Gene Name:
- mhpB
- Uniprot ID:
- P0ABR9
- Molecular weight:
- 34196
Reactions
| 3-(2,3-dihydroxyphenyl)propanoate + O(2) = 2-hydroxy-6-oxonona-2,4-diene-1,9-dioate. |
| (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate + O(2) = 2-hydroxy-6-oxonona-2,4,7-triene-1,9-dioate. |
- General function:
- Involved in 3-phenylpropionate dioxygenase activity
- Specific function:
- Part of the multicomponent 3-phenylpropionate dioxygenase. Converts 3-phenylpropionic acid (PP) and cinnamic acid (CI) into 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol), respectively
- Gene Name:
- hcaF
- Uniprot ID:
- Q47140
- Molecular weight:
- 20579
Reactions
| 3-phenylpropanoate + NADH + O(2) = 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD(+). |
| (2E)-3-phenylprop-2-enoate + NADH + O(2) = (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate + NAD(+). |
- General function:
- Not Available
- Specific function:
- Converts 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol) into 3-(2,3-dihydroxylphenyl)propanoic acid (DHPP) and 2,3-dihydroxicinnamic acid (DHCI), respectively (By similarity). Converts 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol) into 3-(2,3-dihydroxylphenyl)propanoic acid (DHPP) and 2,3-dihydroxicinnamic acid (DHCI), respectively (By similarity).
- Gene Name:
- hcaB
- Uniprot ID:
- P0CI31
- Molecular weight:
- Not Available
Reactions
| 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD(+) = 3-(2,3-dihydroxyphenyl)propanoate + NADH. |
| 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD(+) = 3-(2,3-dihydroxyphenyl)propanoate + NADH. |
| (2E)-3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)prop-2-enoate + NAD(+) = (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate + NADH. |
- General function:
- Involved in 3-(3-hydroxyphenyl)propionate hydroxylase activity
- Specific function:
- Catalyzes the insertion of one atom of molecular oxygen into position 2 of the phenyl ring of 3-(3- hydroxyphenyl)propionate (3-HPP) and hydroxycinnamic acid (3HCI)
- Gene Name:
- mhpA
- Uniprot ID:
- P77397
- Molecular weight:
- 62185
Reactions
| 3-(3-hydroxyphenyl)propanoate + NADH + O(2) = 3-(2,3-dihydroxyphenyl)propanoate + H(2)O + NAD(+). |
| (2E)-3-(3-hydroxyphenyl)prop-2-enoate + NADH + O(2) = (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate + H(2)O + NAD(+). |