Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
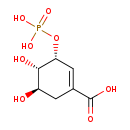
Shikimate 3-phosphate (M2MDB001039)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:32:26 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-09-17 15:42:00 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Shikimate 3-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Shikimate 3-phosphate is a member of the chemical class known as Organophosphate Esters. These are organic compounds containing phosphoric acid ester functional group. Shikimate 3-phosphate is involved in the shikimate pathway. The shikimate pathway enzyme 5-enolpyruvyl shikimate-3-phosphate synthase (EPSP synthase) has received attention in the past because it is the target of the broad-spectrum herbicide glyphosate. (PMID 16225867) The enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) catalyzes the penultimate step of the shikimate pathway and is the target of the broad-spectrum herbicide glyphosate. (PMID 15736934) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C7H8O8P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 251.108 Monoisotopic: 250.997324955 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | QYOJSKGCWNAKGW-PBXRRBTRSA-K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C7H11O8P/c8-4-1-3(7(10)11)2-5(6(4)9)15-16(12,13)14/h2,4-6,8-9H,1H2,(H,10,11)(H2,12,13,14)/p-3/t4-,5-,6+/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (3R,4S,5R)-4,5-dihydroxy-3-(phosphonooxy)cyclohex-1-ene-1-carboxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 3-phosphoshikimic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@@]1(O)CC(=C[C@@]([H])(OP([O-])([O-])=O)[C@@]1([H])O)C([O-])=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as monoalkyl phosphates. These are organic compounds containing a phosphate group that is linked to exactly one alkyl chain. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organic phosphoric acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Phosphate esters | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monoalkyl phosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic homomonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Shikimic acid <> ADP + Hydrogen ion + Shikimate 3-phosphate Phosphoenolpyruvic acid + Shikimate 3-phosphate <> 5-O-(1-Carboxyvinyl)-3-phosphoshikimate + Phosphate Adenosine triphosphate + Shikimic acid <> ADP + Shikimate 3-phosphate Shikimic acid + Adenosine triphosphate > Hydrogen ion + Shikimate 3-phosphate + ADP Phosphoenolpyruvic acid + Shikimate 3-phosphate > Inorganic phosphate + 5-O-(1-Carboxyvinyl)-3-phosphoshikimate Adenosine triphosphate + Shikimic acid > ADP + Shikimate 3-phosphate Shikimic acid + Adenosine triphosphate > Adenosine diphosphate + Hydrogen ion + shikimate 3-phosphate + ADP + Shikimate 3-phosphate shikimate 3-phosphate + Phosphoenolpyruvic acid + Shikimate 3-phosphate > Phosphate + 5-enolpyruvyl-shikimate 3-phosphate Adenosine triphosphate + Shikimic acid <> ADP + Hydrogen ion + Shikimate 3-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in transferase activity, transferring alkyl or aryl (other than methyl) groups
- Specific function:
- Phosphoenolpyruvate + 3-phosphoshikimate = phosphate + 5-O-(1-carboxyvinyl)-3-phosphoshikimate
- Gene Name:
- aroA
- Uniprot ID:
- P0A6D3
- Molecular weight:
- 46095
Reactions
| Phosphoenolpyruvate + 3-phosphoshikimate = phosphate + 5-O-(1-carboxyvinyl)-3-phosphoshikimate. |
- General function:
- Involved in shikimate kinase activity
- Specific function:
- Catalyzes the specific phosphorylation of the 3-hydroxyl group of shikimic acid using ATP as a cosubstrate
- Gene Name:
- aroK
- Uniprot ID:
- P0A6D7
- Molecular weight:
- 19538
Reactions
| ATP + shikimate = ADP + shikimate 3-phosphate. |
- General function:
- Involved in shikimate kinase activity
- Specific function:
- Catalyzes the specific phosphorylation of the 3-hydroxyl group of shikimic acid using ATP as a cosubstrate
- Gene Name:
- aroL
- Uniprot ID:
- P0A6E1
- Molecular weight:
- 19151
Reactions
| ATP + shikimate = ADP + shikimate 3-phosphate. |