Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
S-Ribosyl-L-homocysteine (M2MDB001036)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:32:16 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:19:30 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
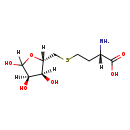
| Name: | S-Ribosyl-L-homocysteine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | S-ribosyl-L-homocysteine is a member of the chemical class known as Alpha Amino Acids and Derivatives. These are amino acids in which the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (alpha carbon). AI-2 is spontaneously derived from 4,5-dihydroxy-2,3-pentanedione that, along with homocysteine, is produced by cleavage of S-adenosylhomocysteine (SAH) and S-ribosylhomocysteine by the Pfs and LuxS enzymes. (PMID 16885435) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C9H17NO6S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 267.299 Monoisotopic: 267.077657971 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | IQFWYNFDWRYSRA-BLELIYKESA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C9H17NO6S/c10-4(8(13)14)1-2-17-3-5-6(11)7(12)9(15)16-5/h4-7,9,11-12,15H,1-3,10H2,(H,13,14)/t4-,5+,6+,7+,9?/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 15912-98-8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-amino-4-({[(2S,3S,4R)-3,4,5-trihydroxyoxolan-2-yl]methyl}sulfanyl)butanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | ribose-5-S-homocysteine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@](N)(CCSC[C@@]1([H])OC([H])(O)[C@]([H])(O)[C@]1([H])O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Pentoses | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | S-Adenosylhomocysteine + Water <> Adenine + S-Ribosyl-L-homocysteine S-Ribosyl-L-homocysteine > 4,5-Dihydroxy-2,3-pentanedione + L-Homocysteine S-Ribosyl-L-homocysteine <> (4S)-4,5-Dihydroxypentan-2,3-dione + L-Homocysteine S-Ribosyl-L-homocysteine + Adenine < S-Adenosylhomocysteine + Water S-Ribosyl-L-homocysteine > L-Homocysteine + (4S)-4,5-Dihydroxypentan-2,3-dione S-Adenosylhomocysteine + Water > S-Ribosyl-L-homocysteine + Adenine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in adenosylhomocysteine nucleosidase activity
- Specific function:
- Catalyzes the irreversible cleavage of the glycosidic bond in both 5'-methylthioadenosine (MTA) and S- adenosylhomocysteine (SAH/AdoHcy) to adenine and the corresponding thioribose, 5'-methylthioribose and S-ribosylhomocysteine, respectively. Can also use 5'-isobutylthioadenosine, 5'-n- butylthioadenosine, S-adenosyl-D-homocysteine, decarboxylated adenosylhomocysteine, deaminated adenosylhomocysteine and S-2-aza- adenosylhomocysteine as substrates
- Gene Name:
- mtnN
- Uniprot ID:
- P0AF12
- Molecular weight:
- 24354
Reactions
| S-adenosyl-L-homocysteine + H(2)O = S-(5-deoxy-D-ribos-5-yl)-L-homocysteine + adenine. |
| S-methyl-5'-thioadenosine + H(2)O = S-methyl-5-thio-D-ribose + adenine. |
- General function:
- Involved in catalytic activity
- Specific function:
- Involved in the synthesis of autoinducer 2 (AI-2) which is secreted by bacteria and is used to communicate both the cell density and the metabolic potential of the environment. The regulation of gene expression in response to changes in cell density is called quorum sensing. Catalyzes the transformation of S-ribosylhomocysteine (RHC) to homocysteine (HC) and 4,5- dihydroxy-2,3-pentadione (DPD)
- Gene Name:
- luxS
- Uniprot ID:
- P45578
- Molecular weight:
- 19416
Reactions
| S-(5-deoxy-D-ribos-5-yl)-L-homocysteine = L-homocysteine + (4S)-4,5-dihydroxypentan-2,3-dione. |