| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:32:13 -0600 |
|---|
| Update Date | 2015-06-03 17:19:29 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | S-(Formylmethyl)glutathione |
|---|
| Description | S-(formylmethyl)glutathione belongs to the class of Peptides. These are compounds containing an amide derived from two or more amino carboxylic acid molecules (the same or different) by formation of a covalent bond from the carbonyl carbon of one to the nitrogen atom of another. (inferred from compound structure) |
|---|
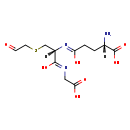
| Structure | |
|---|
| Synonyms: | - (2S)-2-amino-5-(2R)-1-(carboxymethylamino)-1-oxo-3-(2-Oxoethylsulfanyl)propan-2-ylamino-5-oxopentanoate
- (2S)-2-amino-5-(2R)-1-(carboxymethylamino)-1-oxo-3-(2-oxoethylsulfanyl)propan-2-ylamino-5-oxopentanoic acid
- (2S)-2-amino-5-(2R)-1-(carboxymethylamino)-1-oxo-3-(2-Oxoethylsulphanyl)propan-2-ylamino-5-oxopentanoate
- (2S)-2-amino-5-(2R)-1-(carboxymethylamino)-1-oxo-3-(2-Oxoethylsulphanyl)propan-2-ylamino-5-oxopentanoic acid
- L-g-Glutamyl-S-(2-oxoethyl)-L-cysteinylglycine
- L-gamma-Glutamyl-S-(2-oxoethyl)-L-cysteinylglycine
- L-γ-Glutamyl-S-(2-oxoethyl)-L-cysteinylglycine
|
|---|
| Chemical Formula: | C12H19N3O7S |
|---|
| Weight: | Average: 349.36
Monoisotopic: 349.094370667 |
|---|
| InChI Key: | VKLQBAQSSOPXGF-YUMQZZPRSA-N |
|---|
| InChI: | InChI=1S/C12H19N3O7S/c13-7(12(21)22)1-2-9(17)15-8(6-23-4-3-16)11(20)14-5-10(18)19/h3,7-8H,1-2,4-6,13H2,(H,14,20)(H,15,17)(H,18,19)(H,21,22)/t7-,8-/m0/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2S)-2-amino-4-{[(1R)-1-[(carboxymethyl)-C-hydroxycarbonimidoyl]-2-[(2-oxoethyl)sulfanyl]ethyl]-C-hydroxycarbonimidoyl}butanoic acid |
|---|
| Traditional IUPAC Name: | S-(formylmethyl)glutathione |
|---|
| SMILES: | [H][C@](N)(CCC(O)=N[C@@]([H])(CSCC=O)C(O)=NCC(O)=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as oligopeptides. These are organic compounds containing a sequence of between three and ten alpha-amino acids joined by peptide bonds. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Oligopeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-oligopeptide
- Gamma-glutamyl alpha peptide
- Glutamine or derivatives
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Alpha-amino acid amide
- Cysteine or derivatives
- Alpha-amino acid
- N-substituted-alpha-amino acid
- Alpha-amino acid or derivatives
- L-alpha-amino acid
- Dicarboxylic acid or derivatives
- Fatty amide
- Fatty acyl
- Fatty acid
- N-acyl-amine
- Amino acid or derivatives
- Carboxamide group
- Amino acid
- Secondary carboxylic acid amide
- Carboxylic acid
- Thioether
- Sulfenyl compound
- Dialkylthioether
- Primary amine
- Primary aliphatic amine
- Hydrocarbon derivative
- Organic nitrogen compound
- Organic oxygen compound
- Carbonyl group
- Aldehyde
- Amine
- Organic oxide
- Organopnictogen compound
- Organonitrogen compound
- Organooxygen compound
- Organosulfur compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0f6x-9065000000-86d1cba61a3dca3ccc6b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-00di-5211297000-d93bf9d76a2d2fd8d369 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0v59-4189000000-5d5c6497e41130a4bc98 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-9150000000-1a124010fc5e38195c38 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9330000000-0d45185377c925296f38 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00ea-3149000000-2ead9ab4ff107e706425 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00di-9242000000-144fc7fc48f1fe4545b2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9400000000-3beac58433b00fad5b88 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0149000000-53de86e2588eb657c655 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-015i-2952000000-86890d48f98ecf0342c9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-9210000000-bd4daba3840bdfffead2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0592-2189000000-984b1c7e888bfb65e46f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05fu-1690000000-ba60c0a835976ac6512d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-006x-8900000000-1d7b2d35cf7231763801 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|