| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:30:00 -0600 |
|---|
| Update Date | 2015-06-03 17:19:24 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | Formamidopyrimidine nucleoside triphosphate |
|---|
| Description | Formamidopyrimidine nucleoside triphosphate is a member of the chemical class known as Pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. |
|---|
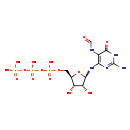
| Structure | |
|---|
| Synonyms: | - (2R,3S,4R,5R)-5-(2-amino-5-formamido-4-oxo-1H-pyrimidin-6-yl)amino-3,4-dihydroxyoxolan-2-ylmethoxy-hydroxyphosphoryl phosphono hydrogen phosphate
- (2R,3S,4R,5R)-5-(2-amino-5-formamido-4-oxo-1H-Pyrimidin-6-yl)amino-3,4-dihydroxyoxolan-2-ylmethoxy-hydroxyphosphoryl phosphono hydrogen phosphoric acid
- Formamidopyrimidine nucleoside triphosphoric acid
- N-(2-amino-5-formamido-6-oxo-3,6-dihydro-4-Pyrimidinyl)-5-O-(hydroxy{hydroxy(phosphonooxy)phosphoryloxy}phosphoryl)-b-D-ribofuranosylamine
- N-(2-Amino-5-formamido-6-oxo-3,6-dihydro-4-pyrimidinyl)-5-O-(hydroxy{hydroxy(phosphonooxy)phosphoryloxy}phosphoryl)-beta-D-ribofuranosylamine
- N-(2-amino-5-formamido-6-oxo-3,6-dihydro-4-Pyrimidinyl)-5-O-(hydroxy{hydroxy(phosphonooxy)phosphoryloxy}phosphoryl)-β-D-ribofuranosylamine
|
|---|
| Chemical Formula: | C10H18N5O15P3 |
|---|
| Weight: | Average: 541.1957
Monoisotopic: 541.001224467 |
|---|
| InChI Key: | NDXMRXXKCCKQQV-UUOKFMHZSA-N |
|---|
| InChI: | InChI=1S/C10H18N5O15P3/c11-10-14-7(4(12-2-16)8(19)15-10)13-9-6(18)5(17)3(28-9)1-27-32(23,24)30-33(25,26)29-31(20,21)22/h2-3,5-6,9,17-18H,1H2,(H,12,16)(H,23,24)(H,25,26)(H2,20,21,22)(H4,11,13,14,15,19)/t3-,5-,6-,9-/m1/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | ({[({[(2R,3S,4R,5R)-5-[(2-amino-5-formamido-6-oxo-1,6-dihydropyrimidin-4-yl)amino]-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)phosphonic acid |
|---|
| Traditional IUPAC Name: | ({[(2R,3S,4R,5R)-5-[(2-amino-5-formamido-6-oxo-1H-pyrimidin-4-yl)amino]-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxyphosphonic acid |
|---|
| SMILES: | NC1=NC(N[C@@H]2O[C@H](COP(O)(=O)OP(O)(=O)OP(O)(O)=O)[C@@H](O)[C@H]2O)=C(NC=O)C(=O)N1 |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- N-arylamide
- Aminopyrimidine
- Pyrimidone
- Secondary aliphatic/aromatic amine
- Monoalkyl phosphate
- Alkyl phosphate
- Hydropyrimidine
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Pyrimidine
- Vinylogous amide
- Tetrahydrofuran
- Heteroaromatic compound
- Amino acid or derivatives
- 1,2-diol
- Carboxamide group
- Secondary carboxylic acid amide
- Secondary alcohol
- Secondary amine
- Azacycle
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Organic nitrogen compound
- Carbonyl group
- Hydrocarbon derivative
- Amine
- Primary amine
- Alcohol
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | - pyrimidine ribonucleoside 5'-triphosphate (CHEBI:27985 )
|
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -4 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | |
|---|
| KEGG Pathways: | |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03dj-6496140000-aad5ec3b4b7957d585bb | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00di-3025009000-fcacb0576bb63da06b26 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dl-0911230000-2740f382fa6662a3aaa2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-0900000000-abf8656c4f6736ab3f89 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-3900000000-98e38554d2fe8f6cb8f0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-02ti-0960050000-9795e309994549a74a15 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9740010000-17fea225289c74811296 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-056r-9610000000-194c76e9de5cd6054fdc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0100190000-10b566c03a24d7543280 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-9636740000-7e8a22838d9f7b49505c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9300100000-92e3fe85b4cbda817fa5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0000090000-5dd4c9b29429f3716be4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01wf-0044390000-a3bec17a6abe49f45c1e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ul0-1497000000-67630f71198823f27b08 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|