| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:29:11 -0600 |
|---|
| Update Date | 2015-06-03 17:19:22 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
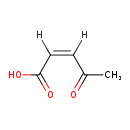
| Name: | cis-Acetylacrylate |
|---|
| Description | Cis-acetylacrylate belongs to the class of Unsaturated Fatty Acids. These are fatty acids whose chain contains at least one CC double bond. (inferred from compound structure) |
|---|
| Structure | |
|---|
| Synonyms: | - Acetylacrylate
- Acetylacrylic acid
- Cis-Acetylacrylic acid
|
|---|
| Chemical Formula: | C5H6O3 |
|---|
| Weight: | Average: 114.0993
Monoisotopic: 114.031694058 |
|---|
| InChI Key: | XGTKSWVCNVUVHG-IHWYPQMZSA-N |
|---|
| InChI: | InChI=1S/C5H6O3/c1-4(6)2-3-5(7)8/h2-3H,1H3,(H,7,8)/b3-2- |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2Z)-4-oxopent-2-enoic acid |
|---|
| Traditional IUPAC Name: | cis-acetylacrylic acid |
|---|
| SMILES: | [H]\C(=C(/[H])C(O)=O)C(C)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Keto acids and derivatives |
|---|
| Sub Class | Short-chain keto acids and derivatives |
|---|
| Direct Parent | Short-chain keto acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Short-chain keto acid
- Fatty acyl
- Unsaturated fatty acid
- Acryloyl-group
- Enone
- Alpha,beta-unsaturated ketone
- Ketone
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxide
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | - Microbial metabolism in diverse environments ec01120
- gamma-Hexachlorocyclohexane degradation ec00361
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-002g-9000000000-2061ae1a8861f42f1c73 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00di-9400000000-f73d0beec9860c695eec | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9100000000-853b506b94b09ffadf54 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ftb-9000000000-12b21331ddede63beb81 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ufr-9000000000-e9848e2a1399185ca489 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-6900000000-f4fc97ecbaa587d81b1b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03xs-9300000000-f9afeb3b5ff5e9af4bb8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0fr2-9000000000-c0b2fd76e44f097e767b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014m-9000000000-1c4c26eebab1a2f95b1c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fr6-9000000000-aa47fe3b96e724c2df4b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-06ea02079a8a17b618db | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-9300000000-1628c355975cd34af232 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-014i-9000000000-818e5c336756d6610c5b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0gb9-9000000000-8840275d4a41857285d2 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Yurtsever D. (2007). Fatty acid methyl ester profiling of Enterococcus and Esherichia coli for microbial source tracking. M.sc. Thesis. Villanova University: U.S.A
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|