Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
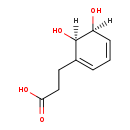
cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol (M2MDB000978)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:29:05 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:19:22 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Cis-3-(carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol (also known as 3-(cis-5,6-Dihydroxycyclohexa-1,3-dien-1-yl)propanoate) is an intermediate invovled in Phenylpropionic acid degradation. It is a substrate for the enzyme 3-phenylpropionate-dihydrodiol/cinnamic acid-dihydrodiol dehydrogenase (hcaB). This enzyme normally converts 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol) into 3-(2,3-dihydroxylphenyl)propanoic acid (DHPP) and 2,3-dihydroxicinnamic acid (DHCI), respectively. Cis-3-(carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol is also a substrate for 3-phenylpropionate/cinnamic acid dioxygenase (hcaE and hcaF), which is also part of the phentylpropionic acid degradatioin pathway. This enzyme catalyzes the reaction 3-phenylpropanoate + NADH + O2 = 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD+ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C9H12O4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 184.1892 Monoisotopic: 184.073558872 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | RKDFGWAXBBGKMR-IONNQARKSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C9H12O4/c10-7-3-1-2-6(9(7)13)4-5-8(11)12/h1-3,7,9-10,13H,4-5H2,(H,11,12)/t7-,9+/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 3-[(5S,6R)-5,6-dihydroxycyclohexa-1,3-dien-1-yl]propanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 3-[(5S,6R)-5,6-dihydroxycyclohexa-1,3-dien-1-yl]propanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | [H][C@]1(O)C=CC=C(CCC(O)=O)[C@@]1([H])O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as 1,2-diols. These are polyols containing an alcohol group at two adjacent positions. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic oxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Alcohols and polyols | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | 1,2-diols | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic homomonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Hydrocinnamic acid + Oxygen + NADH + Hydrogen ion <> cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD <> 3-(2,3-Dihydroxyphenyl)propionic acid + NADH + Hydrogen ion Hydrocinnamic acid + NADH + Oxygen + Hydrogen ion > cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD > Hydrogen ion + 3-(2,3-Dihydroxyphenyl)propionic acid + NADH cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD > 3-(2,3-Dihydroxyphenyl)propanoate + NADH Hydrocinnamic acid + NADH + Oxygen > cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD NADH + Hydrogen ion + Oxygen + Hydrocinnamic acid <> cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD + Trans-2,3-Dihydroxycinnamate cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol + NAD + cis-3-(3-Carboxyethenyl)-3,5-cyclohexadiene-1,2-diol <> 3-(2,3-Dihydroxyphenyl)propionic acid + NADH + Hydrogen ion + Trans-2,3-Dihydroxycinnamate NADH + Oxygen + 3-phenylpropanoate <> NAD + cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol Hydrocinnamic acid + NADH + Oxygen <> NAD + cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in iron ion binding
- Specific function:
- Part of the multicomponent 3-phenylpropionate dioxygenase. Converts 3-phenylpropionic acid (PP) and cinnamic acid (CI) into 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol), respectively
- Gene Name:
- hcaE
- Uniprot ID:
- P0ABR5
- Molecular weight:
- 51109
Reactions
| 3-phenylpropanoate + NADH + O(2) = 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD(+). |
| (2E)-3-phenylprop-2-enoate + NADH + O(2) = (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate + NAD(+). |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Part of the multicomponent 3-phenylpropionate dioxygenase, that converts 3-phenylpropionic acid (PP) and cinnamic acid (CI) into 3-phenylpropionate-dihydrodiol (PP- dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol), respectively. This protein seems to be a 2Fe-2S ferredoxin
- Gene Name:
- hcaC
- Uniprot ID:
- P0ABW0
- Molecular weight:
- 11329
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Part of the multicomponent 3-phenylpropionate dioxygenase, that converts 3-phenylpropionic acid (PP) and cinnamic acid (CI) into 3-phenylpropionate-dihydrodiol (PP- dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol), respectively
- Gene Name:
- hcaD
- Uniprot ID:
- P77650
- Molecular weight:
- 43978
Reactions
| Reduced ferredoxin + NAD(+) = oxidized ferredoxin + NADH. |
- General function:
- Involved in 3-phenylpropionate dioxygenase activity
- Specific function:
- Part of the multicomponent 3-phenylpropionate dioxygenase. Converts 3-phenylpropionic acid (PP) and cinnamic acid (CI) into 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol), respectively
- Gene Name:
- hcaF
- Uniprot ID:
- Q47140
- Molecular weight:
- 20579
Reactions
| 3-phenylpropanoate + NADH + O(2) = 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD(+). |
| (2E)-3-phenylprop-2-enoate + NADH + O(2) = (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate + NAD(+). |
- General function:
- Not Available
- Specific function:
- Converts 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol) into 3-(2,3-dihydroxylphenyl)propanoic acid (DHPP) and 2,3-dihydroxicinnamic acid (DHCI), respectively (By similarity). Converts 3-phenylpropionate-dihydrodiol (PP-dihydrodiol) and cinnamic acid-dihydrodiol (CI-dihydrodiol) into 3-(2,3-dihydroxylphenyl)propanoic acid (DHPP) and 2,3-dihydroxicinnamic acid (DHCI), respectively (By similarity).
- Gene Name:
- hcaB
- Uniprot ID:
- P0CI31
- Molecular weight:
- Not Available
Reactions
| 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD(+) = 3-(2,3-dihydroxyphenyl)propanoate + NADH. |
| 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate + NAD(+) = 3-(2,3-dihydroxyphenyl)propanoate + NADH. |
| (2E)-3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)prop-2-enoate + NAD(+) = (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate + NADH. |