| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:26:48 -0600 |
|---|
| Update Date | 2015-06-03 17:19:17 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
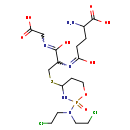
| Name: | 4-Glutathionyl cyclophosphamide |
|---|
| Description | 4-glutathionyl cyclophosphamide belongs to the class of Peptides. These are compounds containing an amide derived from two or more amino carboxylic acid molecules (the same or different) by formation of a covalent bond from the carbonyl carbon of one to the nitrogen atom of another. (inferred from compound structure) |
|---|
| Structure | |
|---|
| Synonyms: | - 2-amino-5-3-2-Bis(2-chloroethyl)amino-2-oxo-1,3,2$l^{5}-oxazaphosphinan-4-ylsulfanyl-1-(carboxymethylamino)-1-oxopropan-2-ylamino-5-oxopentanoate
- 2-amino-5-3-2-bis(2-chloroethyl)amino-2-oxo-1,3,2$l^{5}-oxazaphosphinan-4-ylsulfanyl-1-(carboxymethylamino)-1-oxopropan-2-ylamino-5-oxopentanoic acid
- 2-amino-5-3-2-Bis(2-chloroethyl)amino-2-oxo-1,3,2$l^{5}-oxazaphosphinan-4-ylsulphanyl-1-(carboxymethylamino)-1-oxopropan-2-ylamino-5-oxopentanoate
- 2-amino-5-3-2-Bis(2-chloroethyl)amino-2-oxo-1,3,2$l^{5}-oxazaphosphinan-4-ylsulphanyl-1-(carboxymethylamino)-1-oxopropan-2-ylamino-5-oxopentanoic acid
- g-Glutamyl-S-{2-bis(2-chloroethyl)amino-2-oxido-1,3,2-oxazaphosphinan-4-yl}cysteinylglycine
- Gamma-Glutamyl-S-{2-bis(2-chloroethyl)amino-2-oxido-1,3,2-oxazaphosphinan-4-yl}cysteinylglycine
- γ-Glutamyl-S-{2-bis(2-chloroethyl)amino-2-oxido-1,3,2-oxazaphosphinan-4-yl}cysteinylglycine
|
|---|
| Chemical Formula: | C17H30Cl2N5O8PS |
|---|
| Weight: | Average: 566.394
Monoisotopic: 565.092975577 |
|---|
| InChI Key: | CXEDBYAXQXFDHD-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C17H30Cl2N5O8PS/c18-4-6-24(7-5-19)33(31)23-14(3-8-32-33)34-10-12(16(28)21-9-15(26)27)22-13(25)2-1-11(20)17(29)30/h11-12,14H,1-10,20H2,(H,21,28)(H,22,25)(H,23,31)(H,26,27)(H,29,30) |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | 2-amino-4-{[2-({2-[bis(2-chloroethyl)amino]-2-oxo-1,3,2λ⁵-oxazaphosphinan-4-yl}sulfanyl)-1-[(carboxymethyl)-C-hydroxycarbonimidoyl]ethyl]-C-hydroxycarbonimidoyl}butanoic acid |
|---|
| Traditional IUPAC Name: | C17H30cl2N5O8PS |
|---|
| SMILES: | NC(CCC(O)=NC(CSC1CCOP(=O)(N1)N(CCCl)CCCl)C(O)=NCC(O)=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as oligopeptides. These are organic compounds containing a sequence of between three and ten alpha-amino acids joined by peptide bonds. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Oligopeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-oligopeptide
- Gamma-glutamyl alpha peptide
- Glutamine or derivatives
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Alpha-amino acid amide
- Cysteine or derivatives
- Alpha-amino acid
- N-substituted-alpha-amino acid
- Alpha-amino acid or derivatives
- Nitrogen mustard
- Phosphoric monoester diamide
- Dicarboxylic acid or derivatives
- Fatty amide
- N-acyl-amine
- Organic phosphoric acid derivative
- Oxazaphosphinane
- Fatty acyl
- Organic phosphoric acid amide
- Fatty acid
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Carboxamide group
- Amino acid
- Dialkylthioether
- Sulfenyl compound
- Thioether
- Organoheterocyclic compound
- Carboxylic acid
- Oxacycle
- Azacycle
- Organonitrogen compound
- Alkyl halide
- Primary aliphatic amine
- Organopnictogen compound
- Alkyl chloride
- Primary amine
- Organic oxide
- Carbonyl group
- Organooxygen compound
- Amine
- Hydrocarbon derivative
- Organic oxygen compound
- Organosulfur compound
- Organohalogen compound
- Organic nitrogen compound
- Organochloride
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Not Available |
|---|
| Charge: | -2 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | Not Available |
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0006-9440780000-72a130ffdc0663407ea5 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-06y6-5051192000-85c6673bb5eb4869b5f4 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("4-Glutathionyl cyclophosphamide,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0fk9-3139020000-046b94793b5ffdc91fef | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-006x-9724100000-8aaf34c2e69014649e08 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-9130000000-3b7c56c19d5091ab9e50 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0zgj-0212590000-d3604655628d2a5897dc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9016000000-bf78140826d14fe805c3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9841000000-f940fa82fb8f8898fd89 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-0001960000-dca074efa713b2affcfa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03ki-1036920000-4965fe95e9631cf5a4dd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kr-9005200000-ddb699d034625c7b8bf2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03dj-1000190000-8abecb432b7850e834a8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-024i-4922220000-411f179e15245039ed8a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9820100000-30f2da94e763624e1fab | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|