Structural search and advanced query search is temporarily unavailable. We are working to fix this issue. Thank you for your support and patience.
Phosphoribulosylformimino-AICAR-P (M2MDB000838)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 2.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2012-05-31 14:21:33 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2015-06-03 17:19:04 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Phosphoribulosylformimino-AICAR-P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Phosphoribulosylformimino-aicar-p is a member of the chemical class known as 1-Phosphoribosyl-imidazolecarboxamides. These are organic compounds containing the imidazolecarboxamide linked to a ribose phosphate through a 1-2 bond. PRFAR is catalyzed by imidazole glycerol phosphate syntahse. Imidazole glycerol phosphate (IGP) synthase is a glutamine amidotransferase that catalyzes the formation of IGP and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) from N(1)-[(5'-phosphoribulosyl)formimino]-5-aminoimidazole-4-car boxamide ribonucleotide (PRFAR). (PMID 10733892). 5-((5-phospho-1-deoxyribulos-1-ylamino)methylideneamino)-1-(5-phosphoribosyl)imidazole-4-carboxamide is an intermediate in the histidine biosynthesis pathway. It is a substrate for the enzyme 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase which catalyzes the reaction 1-(5-phosphoribosyl)-5-((5-phosphoribosylamino)methylideneamino)imidazole-4-carboxamide = 5-((5-phospho-1-deoxyribulos-1-ylamino)methylideneamino)-1-(5-phosphoribosyl)imidazole-4-carboxamide. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
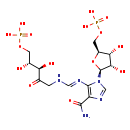
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C15H25N5O15P2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Weight: | Average: 577.331 Monoisotopic: 577.082238179 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | BLKFNHOCHNCLII-IIZOACFYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C15H25N5O15P2/c16-13(26)9-14(18-4-17-1-6(21)10(23)7(22)2-33-36(27,28)29)20(5-19-9)15-12(25)11(24)8(35-15)3-34-37(30,31)32/h4-5,7-8,10-12,15,22-25H,1-3H2,(H2,16,26)(H,17,18)(H2,27,28,29)(H2,30,31,32)/t7-,8+,10+,11+,12+,15+/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(2R,3R)-5-[(E)-N'-{4-carbamoyl-1-[(2S,3S,4R,5S)-3,4-dihydroxy-5-[(phosphonooxy)methyl]oxolan-2-yl]-1H-imidazol-5-yl}methenimidamido]-2,3-dihydroxy-4-oxopentyl]oxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | [(2R,3R)-5-[(E)-N'-{5-carbamoyl-3-[(2S,3S,4R,5S)-3,4-dihydroxy-5-[(phosphonooxy)methyl]oxolan-2-yl]imidazol-4-yl}methenimidamido]-2,3-dihydroxy-4-oxopentyl]oxyphosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | O[C@H](COP(=O)(O)O)[C@@H](O)C(=O)CN\C=N\C1=C(C(=O)N)N=CN1[C@H]1O[C@@H](COP(=O)(O)O)[C@H](O)[C@@H]1O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as 1-ribosyl-imidazolecarboxamides. These are organic compounds containing the imidazole ring linked to a ribose ring through a 1-2 bond. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Imidazole ribonucleosides and ribonucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | 1-ribosyl-imidazolecarboxamides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | 1-ribosyl-imidazolecarboxamides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | L-Glutamine + Phosphoribulosylformimino-AICAR-P > Phosphoribosyl formamidocarboxamide + D-Erythro-imidazole-glycerol-phosphate + L-Glutamate + Hydrogen ion PhosphoribosylformiminoAICAR-phosphate <> Phosphoribulosylformimino-AICAR-P Phosphoribulosylformimino-AICAR-P + L-Glutamine <> D-Erythro-imidazole-glycerol-phosphate + AICAR + L-Glutamate Phosphoribulosylformimino-AICAR-P + L-Glutamine > Hydrogen ion + L-Glutamate + D-Erythro-imidazole-glycerol-phosphate + AICAR PhosphoribosylformiminoAICAR-phosphate > Phosphoribulosylformimino-AICAR-P Phosphoribulosylformimino-AICAR-P + L-Glutamine > L-Glutamic acid + Hydrogen ion + 5-Amino-4-imidazolecarboxyamide + D-Erythro-imidazole-glycerol-phosphate + L-Glutamate PhosphoribosylformiminoAICAR-phosphate <> Phosphoribulosylformimino-AICAR-P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| EcoCyc Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
1. 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase
- General function:
- Involved in catalytic activity
- Specific function:
- 1-(5-phosphoribosyl)-5-((5- phosphoribosylamino)methylideneamino)imidazole-4-carboxamide = 5- ((5-phospho-1-deoxyribulos-1-ylamino)methylideneamino)-1-(5- phosphoribosyl)imidazole-4-carboxamide
- Gene Name:
- hisA
- Uniprot ID:
- P10371
- Molecular weight:
- 26033
Reactions
| 1-(5-phosphoribosyl)-5-((5-phosphoribosylamino)methylideneamino)imidazole-4-carboxamide = 5-((5-phospho-1-deoxyribulos-1-ylamino)methylideneamino)-1-(5-phosphoribosyl)imidazole-4-carboxamide. |
- General function:
- Involved in transferase activity, transferring pentosyl groups
- Specific function:
- IGPS catalyzes the conversion of PRFAR and glutamine to IGP, AICAR and glutamate. The hisH subunit provides the glutamine amidotransferase activity that produces the ammonia necessary to hisF for the synthesis of IGP and AICAR
- Gene Name:
- hisH
- Uniprot ID:
- P60595
- Molecular weight:
- 21653
Reactions
| 5-[(5-phospho-1-deoxyribulos-1-ylamino)methylideneamino]-1-(5-phosphoribosyl)imidazole-4-carboxamide + L-glutamine = imidazole-glycerol phosphate + 5-aminoimidazol-4-carboxamide ribonucleotide + L-glutamate + H(2)O. |
- General function:
- Involved in catalytic activity
- Specific function:
- IGPS catalyzes the conversion of PRFAR and glutamine to IGP, AICAR and glutamate. The hisF subunit catalyzes the cyclization activity that produces IGP and AICAR from PRFAR using the ammonia provided by the hisH subunit
- Gene Name:
- hisF
- Uniprot ID:
- P60664
- Molecular weight:
- 28454
Reactions
| 5-[(5-phospho-1-deoxyribulos-1-ylamino)methylideneamino]-1-(5-phosphoribosyl)imidazole-4-carboxamide + L-glutamine = imidazole-glycerol phosphate + 5-aminoimidazol-4-carboxamide ribonucleotide + L-glutamate + H(2)O. |