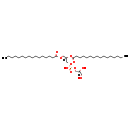| phospholipid biosynthesis (CL(14:0(3-OH)/16:0/16:0/16:0)) | PW001938 | 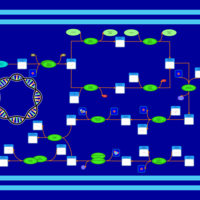 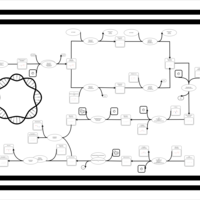 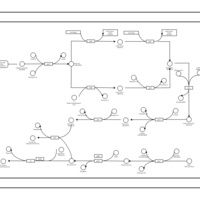 |
| phospholipid biosynthesis (CL(16:0/16:0/16:0/16:0)) | PW002010 |  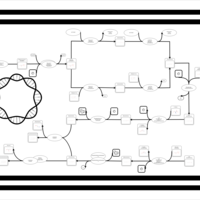 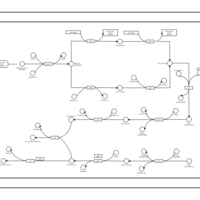 |
| phospholipid biosynthesis (CL(16:1(9Z)/19:0/16:0/16:0)) | PW001961 |  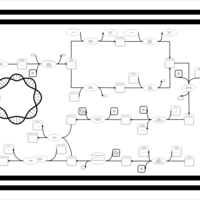 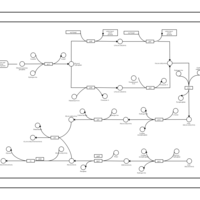 |
| phospholipid biosynthesis (CL(17:0/16:1(9Z)/16:0/16:0)) | PW001964 | 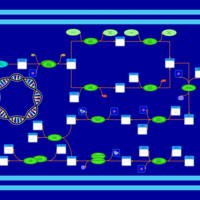 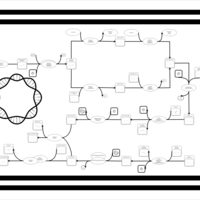 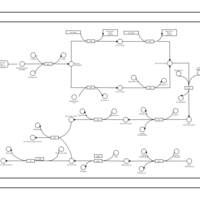 |
| phospholipid biosynthesis (CL(18:1(9Z)/16:0/19:0cycv8c/16:0)) | PW001598 | 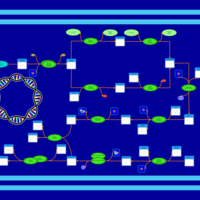   |
| phospholipid biosynthesis (CL(18:1(9Z)/18:1(9Z)/17:0cycw7c/17:0cycw7c)) | PW001679 | 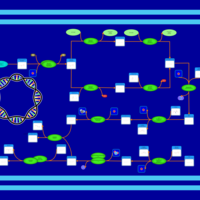 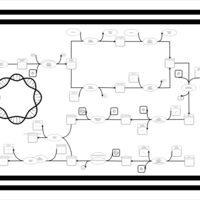 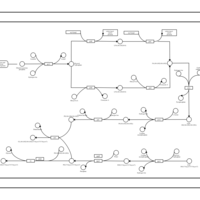 |
| phospholipid biosynthesis (CL(19:0cycv8c/16:0/16:0/14:0)) | PW001323 |  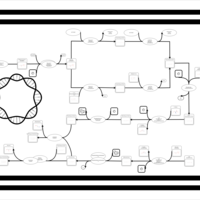 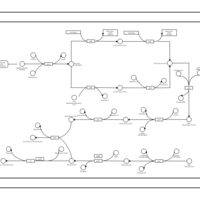 |
| phospholipid biosynthesis (CL(19:0cycv8c/16:0/16:0/17:0cycw7c)) | PW001324 | 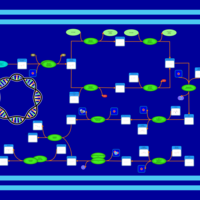   |
| phospholipid biosynthesis (CL(19:0cycv8c/16:0/17:0cycw7c/16:0)) | PW001331 | 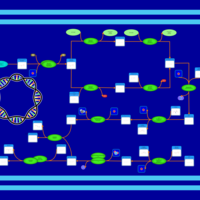   |
| phospholipid biosynthesis CL(15:0cyclo/16:0/16:0/16:0) | PW001100 |   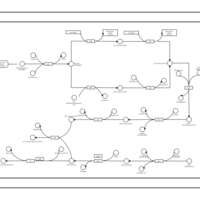 |
| phospholipid biosynthesis CL(15:0cyclo/16:0/16:0/16:1(9Z)) | PW001102 | 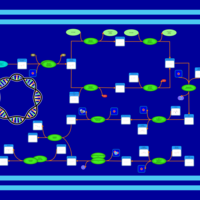 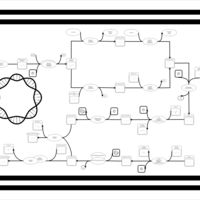 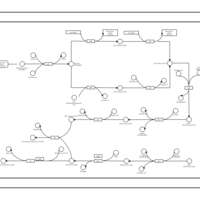 |
| phospholipid biosynthesis CL(15:0cyclo/16:0/16:0/17:0cycw7c) | PW001103 |  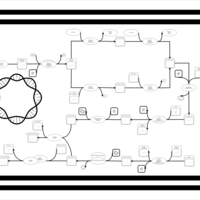 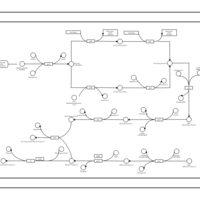 |
| phospholipid biosynthesis CL(15:0cyclo/16:0/16:0/18:1(9Z)) | PW001104 | 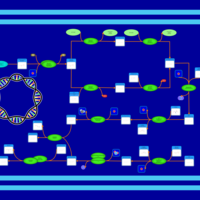 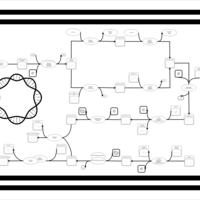 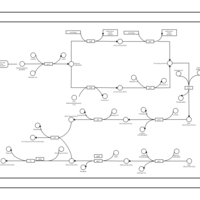 |
| phospholipid biosynthesis CL(15:0cyclo/16:0/16:0/19:0cycv8c) | PW001105 | 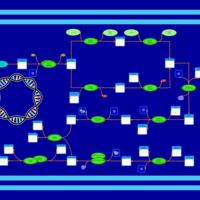  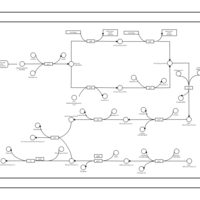 |
| phospholipid biosynthesis CL(15:0cyclo/16:0/16:1(9Z)/16:0) | PW001106 | 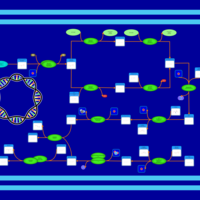 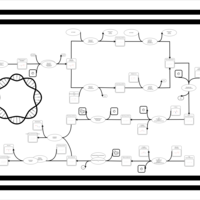 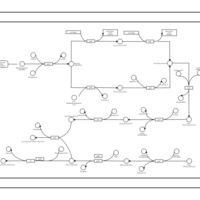 |
| phospholipid biosynthesis CL(15:0cyclo/16:0/17:0cycw7c/16:0) | PW001109 | 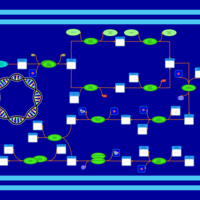  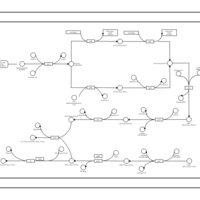 |
| phospholipid biosynthesis CL(15:0cyclo/16:0/18:1(9Z)/16:0) | PW001112 |  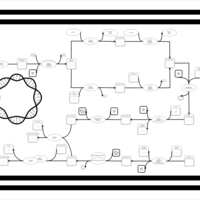 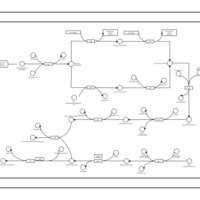 |
| phospholipid biosynthesis CL(15:0cyclo/16:0/19:0cycv8c/16:0) | PW001115 | 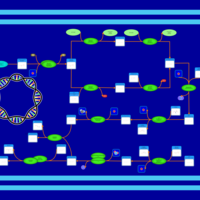 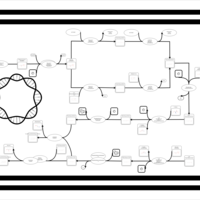 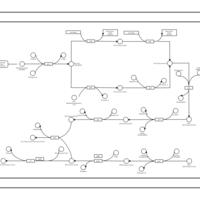 |
| phospholipid biosynthesis CL(15:0cyclo/16:1(9Z)/16:0/16:0) | PW001124 | 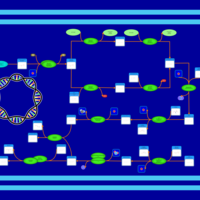 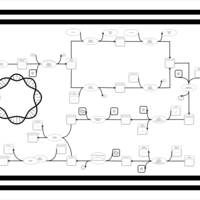  |
| phospholipid biosynthesis CL(15:0cyclo/17:0cycw7c/16:0/16:0) | PW001145 | 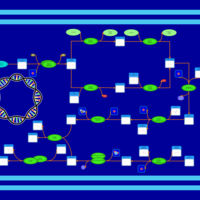 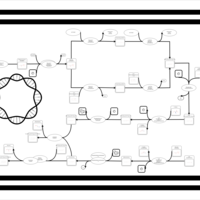 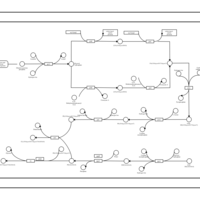 |
| phospholipid biosynthesis CL(15:0cyclo/18:1(9Z)/16:0/16:0) | PW001165 |  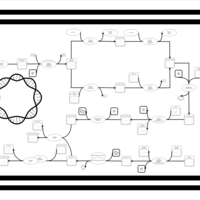  |
| phospholipid biosynthesis CL(15:0cyclo/19:0cycv8c/16:0/16:0) | PW001189 |  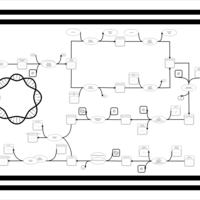 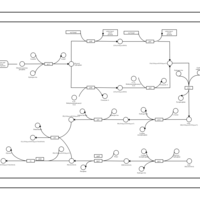 |
| phospholipid biosynthesis CL(16:0/15:0cyclo/16:0/16:0) | PW001226 | 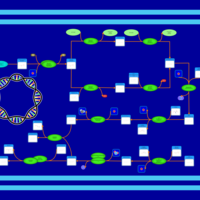 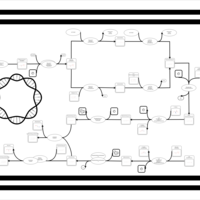 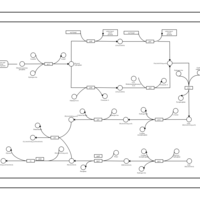 |
| phospholipid biosynthesis CL(16:0/15:0cyclo/16:0/16:1(9Z)) | PW001227 |  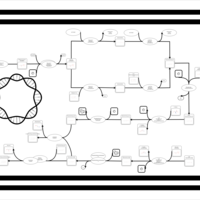  |
| phospholipid biosynthesis CL(16:0/15:0cyclo/16:0/17:0cycw7c) | PW001228 | 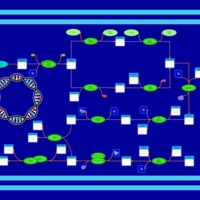  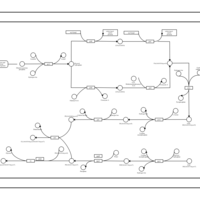 |
| phospholipid biosynthesis CL(16:0/15:0cyclo/16:0/18:1(9Z)) | PW001229 | 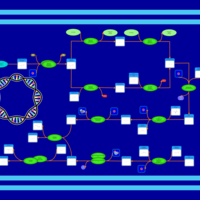 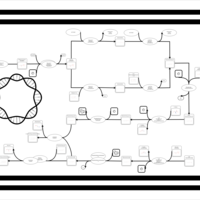 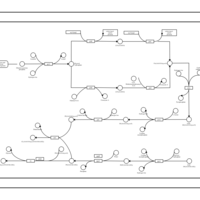 |
| phospholipid biosynthesis CL(16:0/15:0cyclo/16:0/19:0cycv8c) | PW001230 | 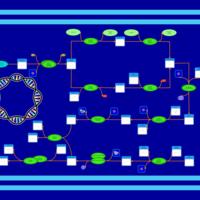  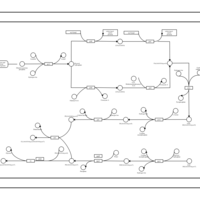 |
| phospholipid biosynthesis CL(16:0/15:0cyclo/16:1(9Z)/16:0) | PW001232 | 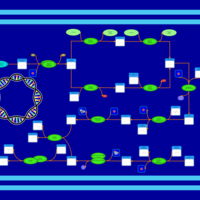 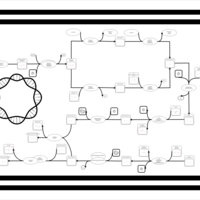 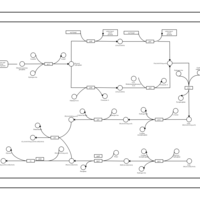 |
| phospholipid biosynthesis CL(16:0/15:0cyclo/17:0cycw7c/16:0) | PW001237 | 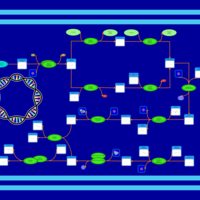  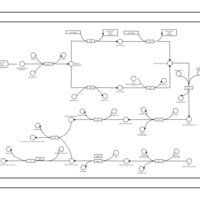 |
| phospholipid biosynthesis CL(16:0/15:0cyclo/18:1(9Z)/16:0) | PW001242 | 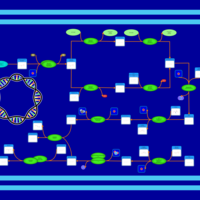 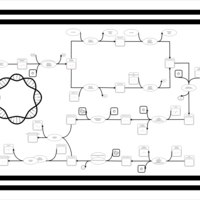 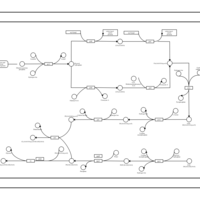 |
| phospholipid biosynthesis CL(16:0/15:0cyclo/19:0cycv8c/16:0) | PW001246 |  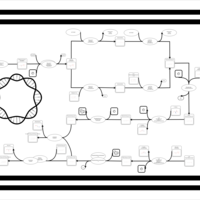  |
| phospholipid biosynthesis CL(16:0/16:0/14:0/14:0) | PW001250 | 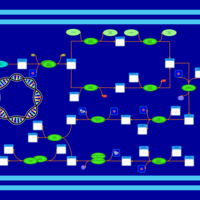 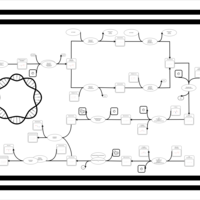 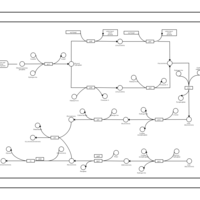 |
| phospholipid biosynthesis CL(16:0/16:0/14:0/16:1(9Z)) | PW001252 | 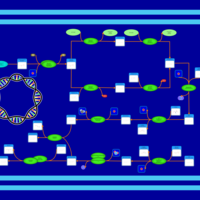 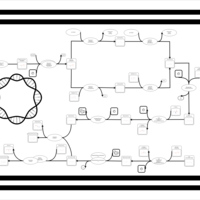 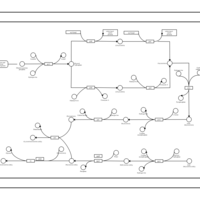 |
| phospholipid biosynthesis CL(16:0/16:0/14:0/17:0cycw7c) | PW001254 |  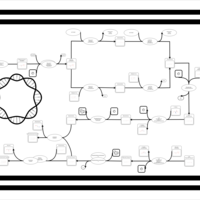 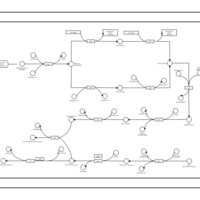 |
| phospholipid biosynthesis CL(16:0/16:0/14:0/18:1(9Z)) | PW001255 |   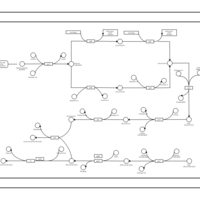 |
| phospholipid biosynthesis CL(16:0/16:0/14:0/19:0cycv8c) | PW001256 |    |
| phospholipid biosynthesis CL(16:0/16:0/16:0/17:0cycw7c) | PW001257 | 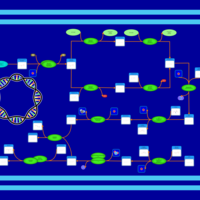   |
| phospholipid biosynthesis CL(16:0/16:0/16:0/18:1(9Z)) | PW001258 | 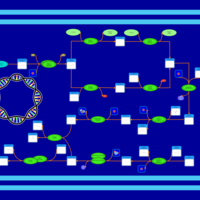   |
| phospholipid biosynthesis CL(16:0/16:0/16:0/19:0cycv8c) | PW001259 | 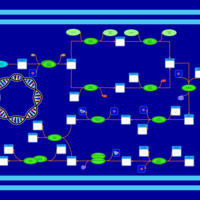 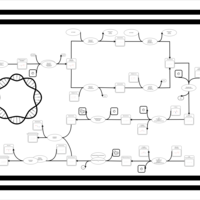 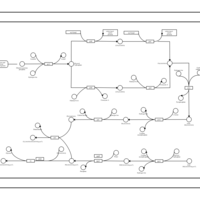 |
| phospholipid biosynthesis CL(16:0/16:0/16:1(9Z)/14:0) | PW001260 |  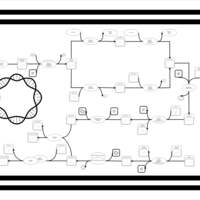  |
| phospholipid biosynthesis CL(16:0/16:0/16:1(9Z)/15:0cyclo) | PW001261 |  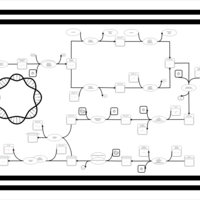 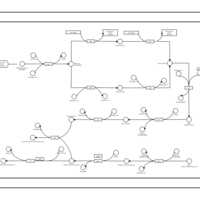 |
| phospholipid biosynthesis CL(16:0/16:0/16:1(9Z)/16:0) | PW001264 |   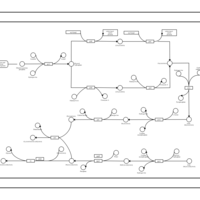 |
| phospholipid biosynthesis CL(16:0/16:0/16:1(9Z)/16:1(9Z)) | PW001265 | 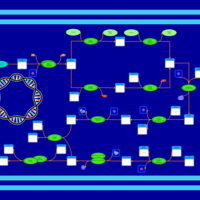 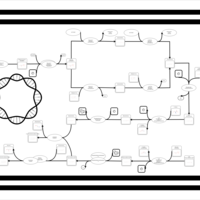  |
| phospholipid biosynthesis CL(16:0/16:0/16:1(9Z)/17:0cycw7c) | PW001267 |    |
| phospholipid biosynthesis CL(16:0/16:0/16:1(9Z)/18:1(9Z)) | PW001268 | 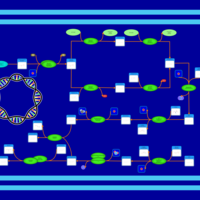   |
| phospholipid biosynthesis CL(16:0/16:0/16:1(9Z)/19:0cycv8c) | PW001269 |  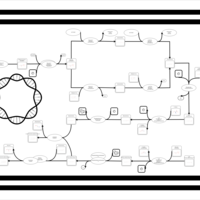 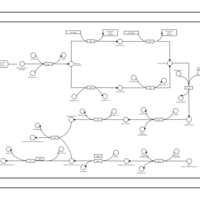 |
| phospholipid biosynthesis CL(16:0/16:0/17:0cycw7c/14:0) | PW001270 | 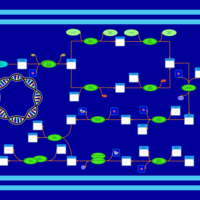   |
| phospholipid biosynthesis CL(16:0/16:0/17:0cycw7c/15:0cyclo) | PW001271 |    |
| phospholipid biosynthesis CL(16:0/16:0/17:0cycw7c/16:0) | PW001272 |    |
| phospholipid biosynthesis CL(16:0/16:0/17:0cycw7c/16:1(9Z)) | PW001273 | 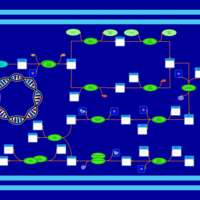 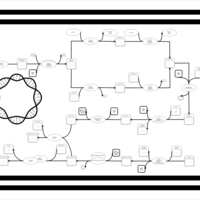 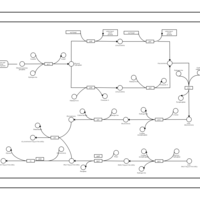 |
| phospholipid biosynthesis CL(16:0/16:0/17:0cycw7c/17:0cycw7c) | PW001274 |  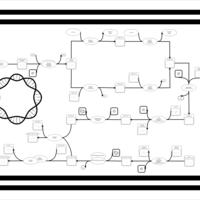  |
| phospholipid biosynthesis CL(16:0/16:0/17:0cycw7c/18:1(9Z)) | PW001277 | 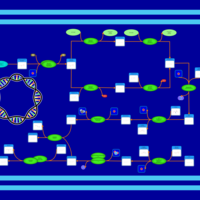 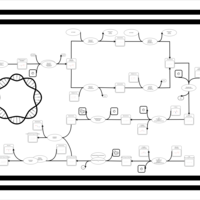 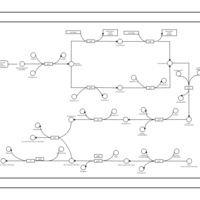 |
| phospholipid biosynthesis CL(16:0/16:0/17:0cycw7c/19:0cycv8c) | PW001278 | 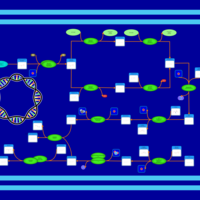 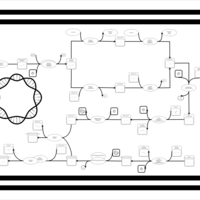 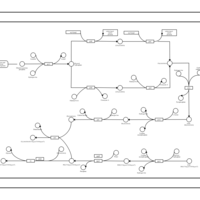 |
| phospholipid biosynthesis CL(16:0/16:0/18:1(9Z)/14:0) | PW001266 | 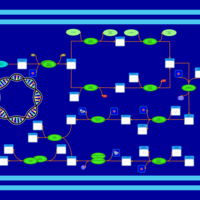 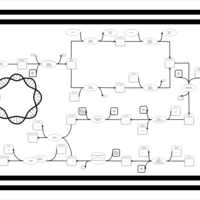 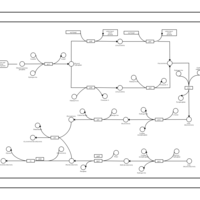 |
| phospholipid biosynthesis CL(16:0/16:0/18:1(9Z)/15:0cyclo) | PW001279 | 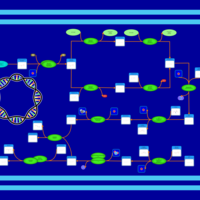   |
| phospholipid biosynthesis CL(16:0/16:0/18:1(9Z)/16:0) | PW001280 | 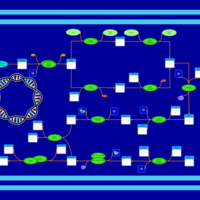 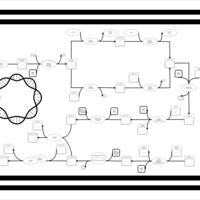 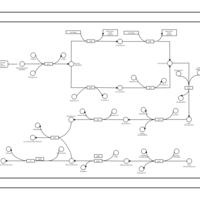 |
| phospholipid biosynthesis CL(16:0/16:0/18:1(9Z)/16:1(9Z)) | PW001281 | 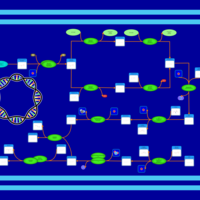 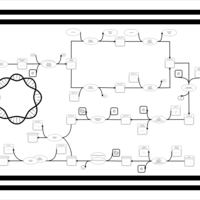 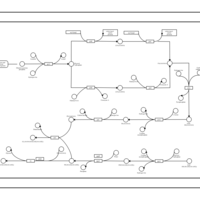 |
| phospholipid biosynthesis CL(16:0/16:0/18:1(9Z)/17:0cycw7c) | PW001282 | 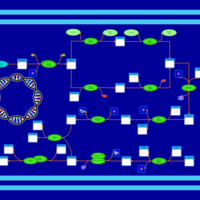  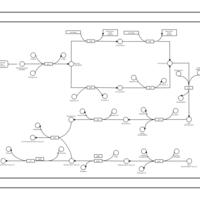 |
| phospholipid biosynthesis CL(16:0/16:0/18:1(9Z)/18:1(9Z)) | PW001283 |    |
| phospholipid biosynthesis CL(16:0/16:0/18:1(9Z)/19:0cycv8c) | PW001284 | 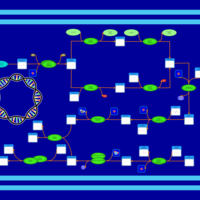 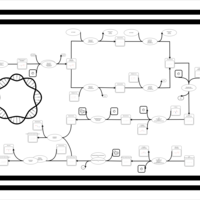  |
| phospholipid biosynthesis CL(16:0/16:0/19:0cycv8c/14:0) | PW001285 |  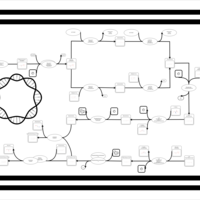  |
| phospholipid biosynthesis CL(16:0/16:0/19:0cycv8c/15:0cyclo) | PW001286 |  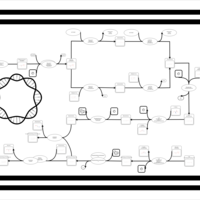  |
| phospholipid biosynthesis CL(16:0/16:0/19:0cycv8c/16:0) | PW001287 |   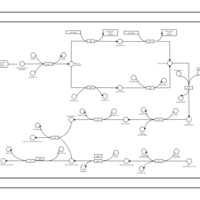 |
| phospholipid biosynthesis CL(16:0/16:0/19:0cycv8c/16:1(9Z)) | PW001288 | 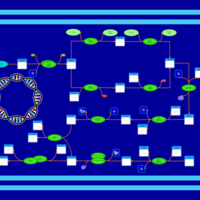  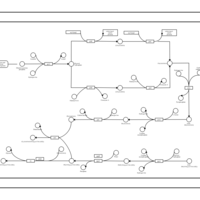 |
| phospholipid biosynthesis CL(16:0/16:0/19:0cycv8c/17:0cycw7c) | PW001289 |  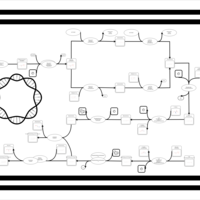 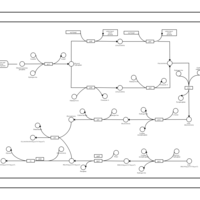 |
| phospholipid biosynthesis CL(16:0/16:0/19:0cycv8c/18:1(9Z)) | PW001290 |  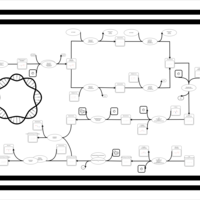 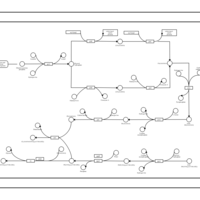 |
| phospholipid biosynthesis CL(16:0/16:1(9Z)/14:0/16:0) | PW001294 | 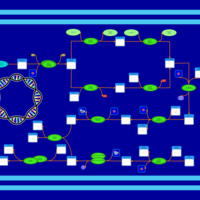   |
| phospholipid biosynthesis CL(16:0/16:1(9Z)/16:0/14:0) | PW001296 | 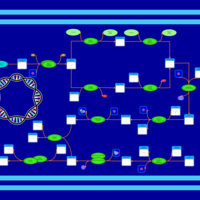   |
| phospholipid biosynthesis CL(16:0/16:1(9Z)/16:0/16:1(9Z)) | PW001297 | 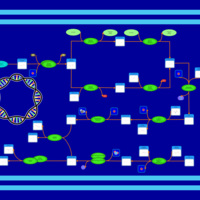   |
| phospholipid biosynthesis CL(16:0/16:1(9Z)/16:0/17:0cycw7c) | PW001303 |  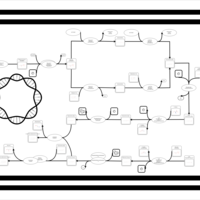 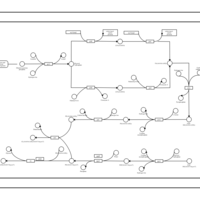 |
| phospholipid biosynthesis CL(16:0/16:1(9Z)/16:0/18:1(9Z)) | PW001305 |  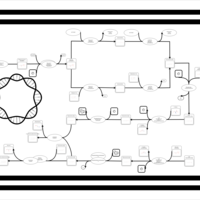 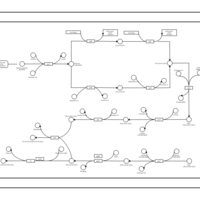 |
| phospholipid biosynthesis CL(16:0/16:1(9Z)/16:0/19:0cycv8c) | PW001306 | 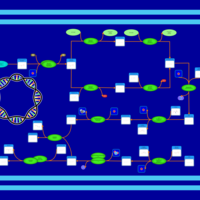 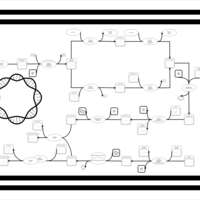 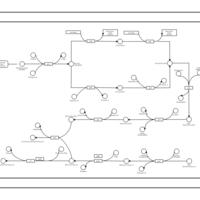 |
| phospholipid biosynthesis CL(16:0/16:1(9Z)/16:1(9Z)/16:0) | PW001308 |  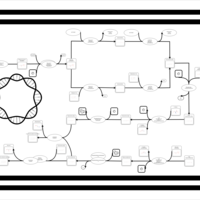  |
| phospholipid biosynthesis CL(16:0/16:1(9Z)/17:0cycw7c/16:0) | PW001315 | 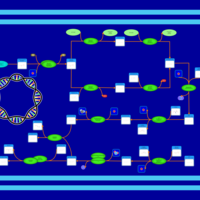   |
| phospholipid biosynthesis CL(16:0/16:1(9Z)/18:1(9Z)/16:0) | PW001328 | 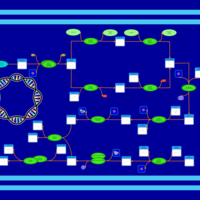 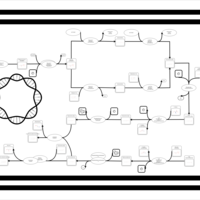  |
| phospholipid biosynthesis CL(16:0/16:1(9Z)/19:0cycv8c/16:0) | PW001335 | 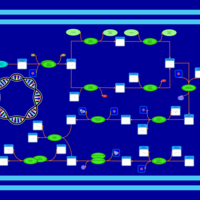 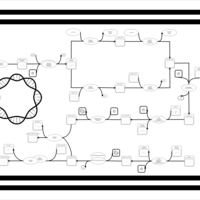  |
| phospholipid biosynthesis CL(16:0/17:0cycw7c/14:0/16:0) | PW001338 | 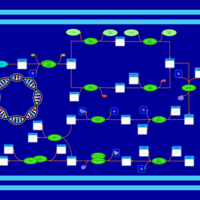   |
| phospholipid biosynthesis CL(16:0/17:0cycw7c/16:0/14:0) | PW001342 | 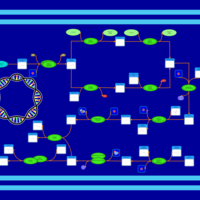   |
| phospholipid biosynthesis CL(16:0/17:0cycw7c/16:0/17:0cycw7c) | PW001343 | 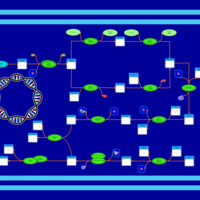 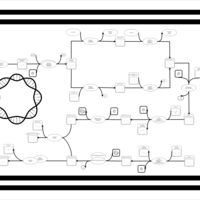 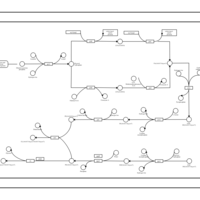 |
| phospholipid biosynthesis CL(16:0/18:1(9Z)/14:0/16:0) | PW001356 | 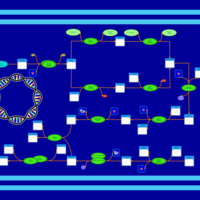  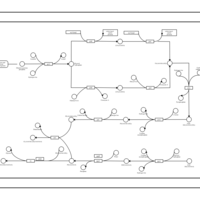 |
| phospholipid biosynthesis CL(16:0/18:1(9Z)/16:0/14:0) | PW001358 | 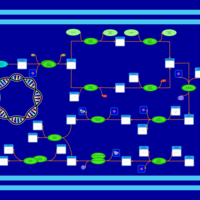 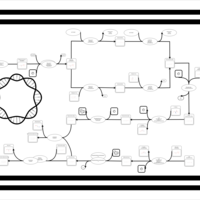 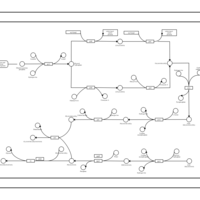 |
| phospholipid biosynthesis CL(16:0/18:1(9Z)/16:0/17:0cycw7c) | PW001359 | 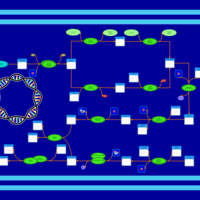 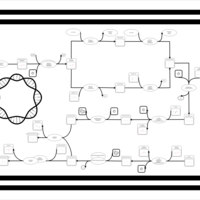  |
| phospholipid biosynthesis CL(16:0/18:1(9Z)/16:0/18:1(9Z)) | PW001360 | 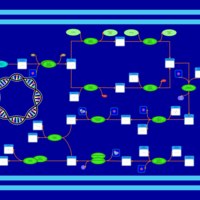 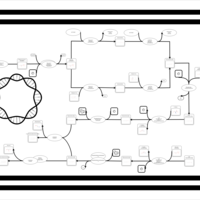  |
| phospholipid biosynthesis CL(16:0/18:1(9Z)/16:0/19:0cycv8c) | PW001361 | 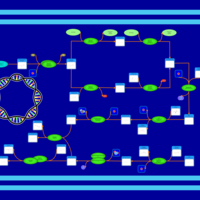 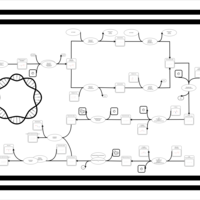  |
| phospholipid biosynthesis CL(16:0/18:1(9Z)/17:0cycw7c/16:0) | PW001364 |  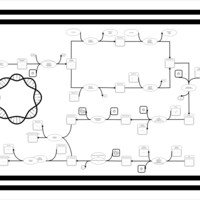  |
| phospholipid biosynthesis CL(16:0/18:1(9Z)/18:1(9Z)/16:0) | PW001368 | 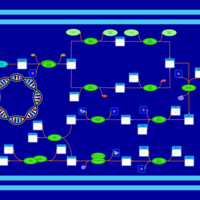 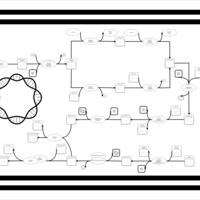 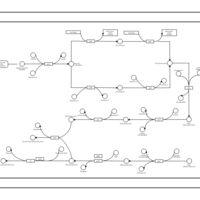 |
| phospholipid biosynthesis CL(16:0/18:1(9Z)/19:0cycv8c/16:0) | PW001379 | 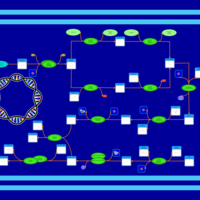   |
| phospholipid biosynthesis CL(16:0/19:0cycv8c/14:0/16:0) | PW001389 | 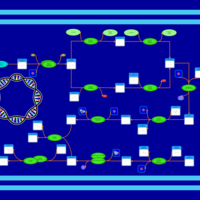 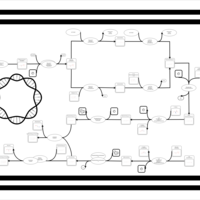 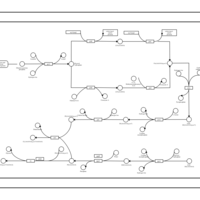 |
| phospholipid biosynthesis CL(16:0/19:0cycv8c/16:0/14:0) | PW001391 | 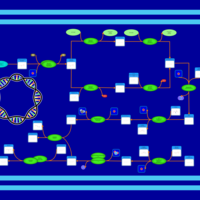 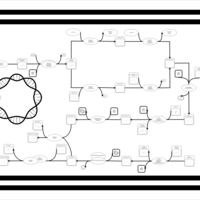 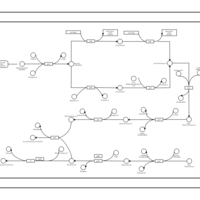 |
| phospholipid biosynthesis CL(16:0/19:0cycv8c/16:0/17:0cycw7c) | PW001392 | 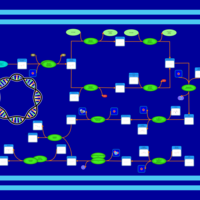 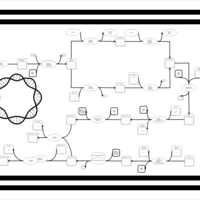 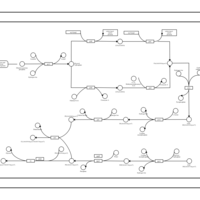 |
| phospholipid biosynthesis CL(16:0/19:0cycv8c/17:0cycw7c/16:0) | PW001399 | 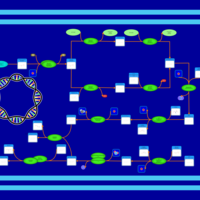 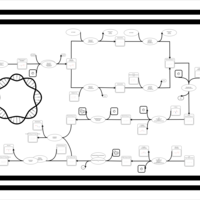  |
| phospholipid biosynthesis CL(16:1(9Z)/16:0/14:0/16:0) | PW001471 |   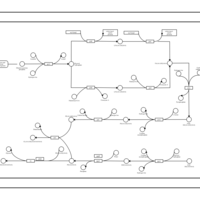 |
| phospholipid biosynthesis CL(16:1(9Z)/16:0/16:0/14:0) | PW001477 |   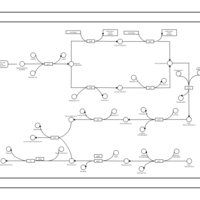 |
| phospholipid biosynthesis CL(16:1(9Z)/16:0/16:0/17:0cycw7c) | PW001478 | 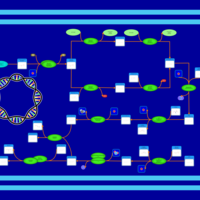 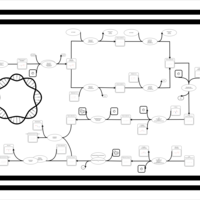 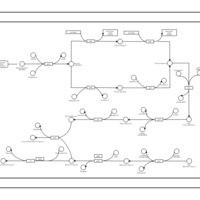 |
| phospholipid biosynthesis CL(16:1(9Z)/16:0/16:0/18:1(9Z)) | PW001479 |   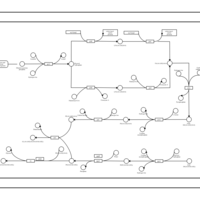 |
| phospholipid biosynthesis CL(16:1(9Z)/16:0/16:0/19:0cycv8c) | PW001480 |  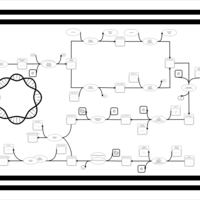 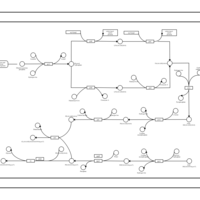 |
| phospholipid biosynthesis CL(16:1(9Z)/16:0/16:1(9Z)/16:0) | PW001486 | 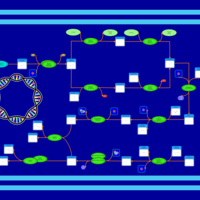 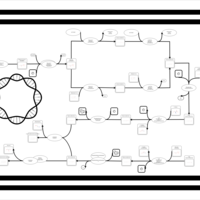  |
| phospholipid biosynthesis CL(16:1(9Z)/16:0/17:0cycw7c/16:0) | PW001491 |   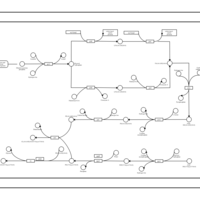 |
| phospholipid biosynthesis CL(16:1(9Z)/16:0/18:1(9Z)/16:0) | PW001501 |    |
| phospholipid biosynthesis CL(16:1(9Z)/16:0/19:0cycv8c/16:0) | PW001505 |    |
| phospholipid biosynthesis CL(16:1(9Z)/16:0/19:0cycv8c/19:0cycv8c) | PW001509 |   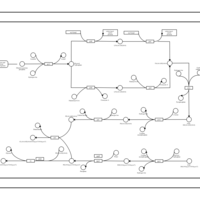 |
| phospholipid biosynthesis CL(17:0cycw7c/16:0/14:0/16:0) | PW001663 | 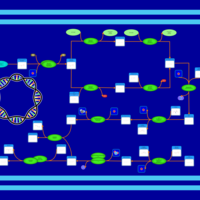 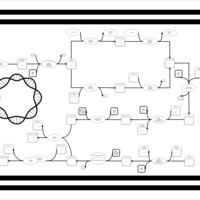 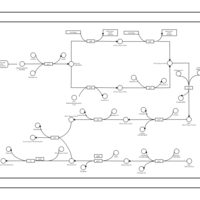 |
| phospholipid biosynthesis CL(17:0cycw7c/16:0/16:0/14:0) | PW001675 | 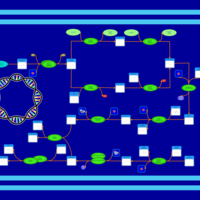 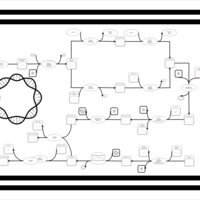 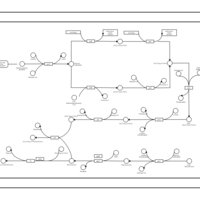 |
| phospholipid biosynthesis CL(17:0cycw7c/16:0/16:0/17:0cycw7c) | PW001676 | 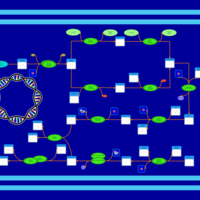 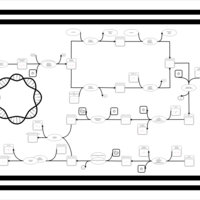 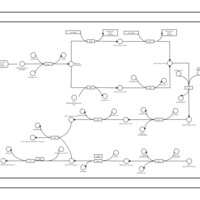 |
| phospholipid biosynthesis CL(17:0cycw7c/16:0/17:0cycw7c/16:0) | PW001681 | 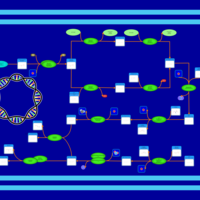 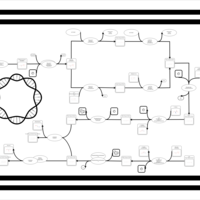  |