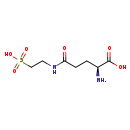| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:07:29 -0600 |
|---|
| Update Date | 2015-06-03 15:54:50 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
| Name: | 5-L-Glutamyl-taurine |
|---|
| Description | 5-L-Glutamyl-taurine is an intermediate in taurine and hypotaurine metabolism. 5-L-Glutamyl-taurine is produced from Taurine via the enzyme gamma-glutamyltranspeptidase (EC 2.3.2.2). |
|---|
| Structure | |
|---|
| Synonyms: | |
|---|
| Chemical Formula: | C7H14N2O6S |
|---|
| Weight: | Average: 254.261
Monoisotopic: 254.05725688 |
|---|
| InChI Key: | WGXUDTHMEITUBO-YFKPBYRVSA-N |
|---|
| InChI: | InChI=1S/C7H14N2O6S/c8-5(7(11)12)1-2-6(10)9-3-4-16(13,14)15/h5H,1-4,8H2,(H,9,10)(H,11,12)(H,13,14,15)/t5-/m0/s1 |
|---|
| CAS number: | Not Available |
|---|
| IUPAC Name: | (2S)-2-amino-4-[(2-sulfoethyl)carbamoyl]butanoic acid |
|---|
| Traditional IUPAC Name: | (2S)-2-amino-4-[(2-sulfoethyl)carbamoyl]butanoic acid |
|---|
| SMILES: | N[C@@H](CCC(=O)NCCS(O)(=O)=O)C(O)=O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glutamine and derivatives. Glutamine and derivatives are compounds containing glutamine or a derivative thereof resulting from reaction of glutamine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Glutamine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glutamine or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Fatty amide
- N-acyl-amine
- Fatty acid
- Fatty acyl
- Organic sulfonic acid or derivatives
- Organosulfonic acid or derivatives
- Organosulfonic acid
- Sulfonyl
- Alkanesulfonic acid
- Amino acid
- Carboxamide group
- Secondary carboxylic acid amide
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Primary amine
- Primary aliphatic amine
- Organic oxygen compound
- Carbonyl group
- Organic nitrogen compound
- Organic oxide
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | -1 |
|---|
| Melting point: | Not Available |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | - Taurine and hypotaurine metabolism ec00430
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0536-9620000000-a538eb9e6c595cef5a4c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-05fu-9551000000-f0e6775174afe08371f3 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-1690000000-5a8d4b6641b218cf8506 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6r-2920000000-360e2479ab5fa4d3ed08 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9400000000-bf155960a2593c2884d1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-2290000000-148d482f4c7749eb7d10 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ff9-7970000000-1c9d1a98595ca5fdde6c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-008c-9200000000-e084fb39cebaaa6d3226 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-055r-0690000000-5751afe071b994d33ca7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9700000000-bd8c4ab66c89ab08c075 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-053r-9300000000-d9853bca074f4454ba2a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0090000000-fb89fab28b92fcc47cb0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fki-4690000000-644bf95f8c720f6682bc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9500000000-457385fb84f7a5c60d8c | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available | View in JSpectraViewer |
|---|
|
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Not Available |
|---|
| Links |
|---|
| External Links: | |
|---|