| Record Information |
|---|
| Version | 2.0 |
|---|
| Creation Date | 2012-05-31 14:04:52 -0600 |
|---|
| Update Date | 2015-09-13 12:56:14 -0600 |
|---|
| Secondary Accession Numbers | |
|---|
| Identification |
|---|
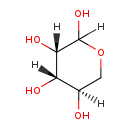
| Name: | D-Lyxose |
|---|
| Description | D-lyxose is a member of the chemical class known as Hexoses. These are monosaccharides in which the sugar unit is a hexose. Lyxose is an aldopentose - a monosaccharide containing five carbon atoms, and including an aldehyde functional group. It has chemical formula C5H10O5. Lyxose occurs only rarely in nature, for example, as a component of bacterial glycolipids (WikiPedia) |
|---|
| Structure | |
|---|
| Synonyms: | - α-D-lyxose
- (+)-Xylose
- 2,3,4,5-Tetrahydroxypentanal
- a-D-Lyxose
- Alpha-D-Lyxose
- D-Lyxose
- DL-Xylose
- L(+)-Xylose
- L-Lyxose
- Lyxose
- Pectin
- Pectin sugar
- Pectinose
- Pentose
- Trobicin
- α-D-Lyxose
|
|---|
| Chemical Formula: | C5H10O5 |
|---|
| Weight: | Average: 150.1299
Monoisotopic: 150.05282343 |
|---|
| InChI Key: | SRBFZHDQGSBBOR-AGQMPKSLSA-N |
|---|
| InChI: | InChI=1S/C5H10O5/c6-2-1-10-5(9)4(8)3(2)7/h2-9H,1H2/t2-,3+,4+,5?/m1/s1 |
|---|
| CAS number: | 1114-34-7 |
|---|
| IUPAC Name: | (3S,4S,5R)-oxane-2,3,4,5-tetrol |
|---|
| Traditional IUPAC Name: | (3S,4S,5R)-oxane-2,3,4,5-tetrol |
|---|
| SMILES: | [H][C@@]1(O)COC([H])(O)[C@@]([H])(O)[C@@]1([H])O |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pentoses. These are monosaccharides in which the carbohydrate moiety contains five carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose monosaccharide
- Oxane
- Secondary alcohol
- Hemiacetal
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Hydrocarbon derivative
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Physical Properties |
|---|
| State: | Solid |
|---|
| Charge: | 0 |
|---|
| Melting point: | 106.5 °C |
|---|
| Experimental Properties: | |
|---|
| Predicted Properties | |
|---|
| Biological Properties |
|---|
| Cellular Locations: | Cytoplasm |
|---|
| Reactions: | |
|---|
| SMPDB Pathways: | Not Available |
|---|
| KEGG Pathways: | - Pentose and glucuronate interconversions ec00040
|
|---|
| EcoCyc Pathways: | Not Available |
|---|
| Concentrations |
|---|
| Not Available |
|---|
| Spectra |
|---|
| Spectra: | |
|---|
| References |
|---|
| References: | - Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| Links |
|---|
| External Links: | |
|---|